Team:USTC Software/demo
From 2010.igem.org
(→modelling and simulation) |
(→modelling and simulation) |
||
| Line 15: | Line 15: | ||
Model written in '''SBML''' format is available here. The dynamic curve of E.coli replication is plotted by time course simulation: | Model written in '''SBML''' format is available here. The dynamic curve of E.coli replication is plotted by time course simulation: | ||
| + | ===the XML=== | ||
| - | + | <?xml version="1.0" encoding="UTF-8"?> | |
| - | + | <sbml xmlns="http://www.sbml.org/sbml/level2/version4" level="2" version="4"> | |
| - | + | <model id="iGameModel"> | |
| - | + | <listOfUnitDefinitions> | |
| - | + | <unitDefinition id="per_second"> | |
| - | + | <listOfUnits> | |
| - | + | <unit kind="second" exponent="-1"/> | |
| - | + | </listOfUnits> | |
| - | + | </unitDefinition> | |
| - | + | <unitDefinition id="mole_per_litre"> | |
| - | + | <listOfUnits> | |
| - | + | <unit kind="mole"/> | |
| - | + | <unit kind="litre" exponent="-1"/> | |
| - | + | </listOfUnits> | |
| - | + | </unitDefinition> | |
| - | + | </listOfUnitDefinitions> | |
| - | + | <listOfCompartments> | |
| - | + | <compartment id="Flask" size="0.4"/> | |
| - | + | <compartment id="E_coli" size="1e-14" outside="Flask"/> | |
| + | </listOfCompartments> | ||
| + | <listOfSpecies> | ||
| + | <species id="sPecIes0" compartment="Flask" initialConcentration="1e-20" substanceUnits="mole"/> | ||
| + | </listOfSpecies> | ||
| + | <listOfReactions> | ||
| + | <reaction id="rEactIon0" name="reproduction of Ecoli" reversible="false" fast="false"> | ||
| + | <listOfProducts> | ||
| + | <speciesReference species="sPecIes0"/> | ||
| + | </listOfProducts> | ||
| + | <listOfModifiers> | ||
| + | <modifierSpeciesReference species="sPecIes0"/> | ||
| + | </listOfModifiers> | ||
| + | <kineticLaw> | ||
| + | <math xmlns="http://www.w3.org/1998/Math/MathML"> | ||
| + | <apply> | ||
| + | <times/> | ||
| + | <ci> kgrowth </ci> | ||
| + | <apply> | ||
| + | <minus/> | ||
| + | <cn type="integer"> 1 </cn> | ||
| + | <apply> | ||
| + | <divide/> | ||
| + | <ci> sPecIes0 </ci> | ||
| + | <ci> maxc </ci> | ||
| + | </apply> | ||
| + | </apply> | ||
| + | <ci> sPecIes0 </ci> | ||
| + | <ci> Flask </ci> | ||
| + | </apply> | ||
| + | </math> | ||
| + | <listOfParameters> | ||
| + | <parameter id="kgrowth" name="growth rate" value="0.0005766" units="per_second"/> | ||
| + | <parameter id="maxc" name="max concentration" value="1.66e-12" units="mole_per_litre"/> | ||
| + | </listOfParameters> | ||
| + | </kineticLaw> | ||
| + | </reaction> | ||
| + | </listOfReactions> | ||
| + | </model> | ||
| + | </sbml> | ||
Revision as of 03:26, 14 October 2010
Using C-N Model we can model systems with great complexity
The simplest example with only E.coli cell
introduction
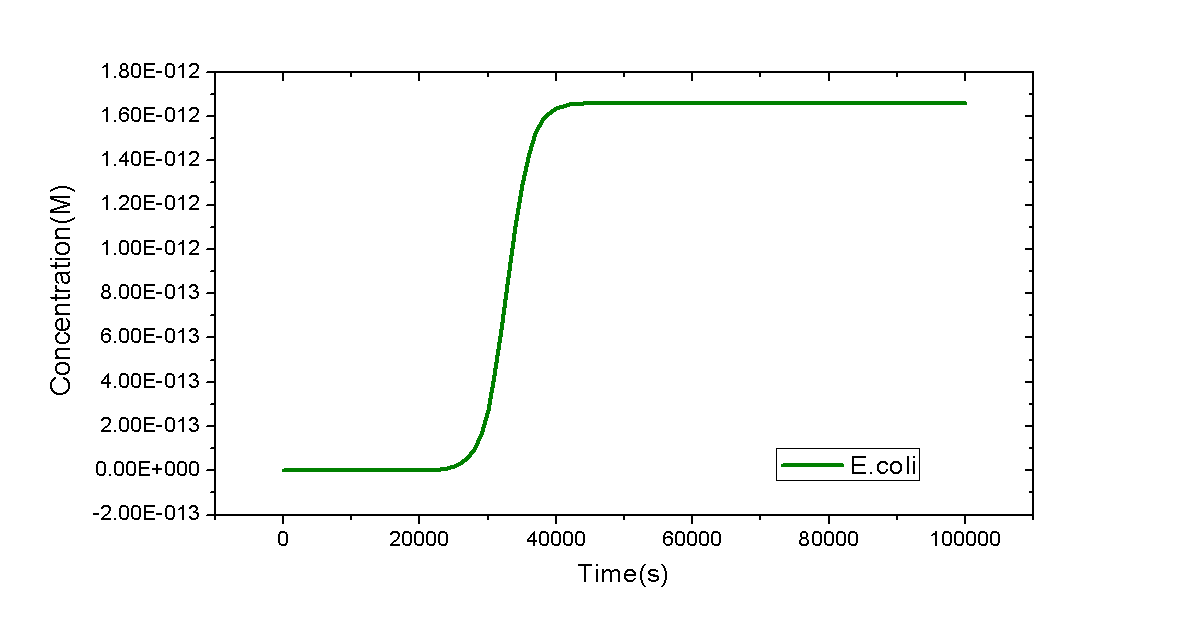
As the simplest example, a model of a flask with only E.coli cells inside will be shown. Only one reaction will be considered: the replication of E.coli. However, it is not trivial. MoDeL enables users to add such an auto-catalytic replication reaction easily and conveniently. The minimal database used for this model could be download here.
database construction
We only provide key points regarded with construction of this minimal database. To add the auto-catalytic reaction in the Reaction container, a species with only part E.coli is required. It has the simplest Chain-Node model format: only one chain with one part and no trees. The auto-catalytic reaction has one modifier and one product and they are both referred to E.coli defined in Species container. Since they are compartment-type species (species representing a compartment), attribute itself of compartmentLabel node in modifiers and products definitions should be set the same with label of the compartment they represent in the compartments definition. It ensures that the product and the modifier are the same, avoiding wrong mismatch of the product which is different with the modifier. Since number of E.coli cells will reach a stable level in a long time course, we use <math>k_{g}(1-C_{E.coli}/C_{max})C_{E.coli}V_{Flask}</math> as the reaction rate, where <math>k_{g}</math> is the growth rate of E.coli, <math>V_{Flask}</math> represents the size of E.coli, and <math>C_{E.coli}</math> and <math> C_{max}</math> are concentration of E.coli and its max concentration in the flask, respectively. The negative sign in the rate equation indicates self-repression of E.coli cells.
modelling and simulation
Model written in SBML format is available here. The dynamic curve of E.coli replication is plotted by time course simulation:
the XML
<?xml version="1.0" encoding="UTF-8"?> <sbml xmlns="http://www.sbml.org/sbml/level2/version4" level="2" version="4">
<model id="iGameModel">
<listOfUnitDefinitions> <unitDefinition id="per_second"> <listOfUnits> <unit kind="second" exponent="-1"/> </listOfUnits> </unitDefinition> <unitDefinition id="mole_per_litre"> <listOfUnits> <unit kind="mole"/> <unit kind="litre" exponent="-1"/> </listOfUnits> </unitDefinition> </listOfUnitDefinitions> <listOfCompartments> <compartment id="Flask" size="0.4"/> <compartment id="E_coli" size="1e-14" outside="Flask"/> </listOfCompartments> <listOfSpecies> <species id="sPecIes0" compartment="Flask" initialConcentration="1e-20" substanceUnits="mole"/> </listOfSpecies> <listOfReactions> <reaction id="rEactIon0" name="reproduction of Ecoli" reversible="false" fast="false"> <listOfProducts> <speciesReference species="sPecIes0"/> </listOfProducts> <listOfModifiers> <modifierSpeciesReference species="sPecIes0"/> </listOfModifiers> <kineticLaw> <math xmlns="http://www.w3.org/1998/Math/MathML"> <apply> <times/> <ci> kgrowth </ci> <apply> <minus/> <cn type="integer"> 1 </cn> <apply> <divide/> <ci> sPecIes0 </ci> <ci> maxc </ci> </apply> </apply> <ci> sPecIes0 </ci> <ci> Flask </ci> </apply> </math> <listOfParameters> <parameter id="kgrowth" name="growth rate" value="0.0005766" units="per_second"/> <parameter id="maxc" name="max concentration" value="1.66e-12" units="mole_per_litre"/> </listOfParameters> </kineticLaw> </reaction> </listOfReactions>
</model>
</sbml>
=============================
Basic example with pLac-LacI repression system
introduction
MoDeL provides poweful supports for description of interactions between two species. No interactions exist in the previous example. In this example, we will construct a plasmid backbone with promoter, rbs, coding sequence and terminator inserted and transform it into E.coli. LacI repressor are produced after two steps of transcription and translation. It binds with itself to form a dimer and then a tetramer, which could bind with promoter LacI on the plasmid to repress the process of transcription. Besides, replication of E.coli cells, degradation of proteins and mRNAs, as well as dilution due to replication of cells, are also considered.
 "
"