Team:TU Delft/19 July 2010 content
From 2010.igem.org
(→Emulsifier) |
(→Emulsifier) |
||
| Line 88: | Line 88: | ||
Lane description: | Lane description: | ||
| - | {| | + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" |
|'''#''' | |'''#''' | ||
|'''Description''' | |'''Description''' | ||
| Line 110: | Line 110: | ||
'''Ligation''' | '''Ligation''' | ||
| - | + | The digestion products were [[Team:TU_Delft/protocols/ligation|ligated]] over night: | |
| + | |||
| + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Ligation reaction''' | ||
| + | |- | ||
| + | |1 | ||
| + | |5 μL Cut plasmid with B0032 + 10 μL PCR Product of R0011 | ||
| + | |} | ||
Revision as of 14:14, 19 July 2010
Lab work
Alkane degradation
Biobricks in production:
| # | Digestion reaction | Used Buffer | Needed fragment |
| 1 | 2 μg J61100 + EcoRI + SpeI | NEBuffer 2 | ‘E – J61100 – S’ |
| 2 | 2 μg J61100 + EcoRI + SpeI | NEBuffer 2 | ‘E – J61100 – S’ |
| 3 | 1 μg rubR + EcoRI + SpeI | NEBuffer 2 | ‘E – rubR– S’ |
| 4 | 1 μg J61101 + EcoRI + SpeI | NEBuffer 2 | ‘E – J61101– S’ |
| 5 | 1 μg J61107 + EcoRI + SpeI | NEBuffer 2 | ‘E – J61107– S’ |
| 6 | 1μg alkB2 + Xbal + PstI | NEBuffer 2 | ‘X – alkB2 – P’ |
| 7 | 1 μg rubA3 + Xbal + PstI | NEBuffer 2 | ‘X – rubA3 – P’ |
| 8 | 1 μg rubA4 + Xbal + PstI | NEBuffer 2 | ‘X – rubA4 – P’ |
| 9 | 1 μg B0015 + Xbal + PstI | NEBuffer 2 | ‘X – B0015 – P’ |
| 10 | 1 μg ladA + Xbal + PstI | NEBuffer 2 | ‘X – ladA – P’ |
| 11 | 1 μg ADH + Xbal + PstI | NEBuffer 2 | ‘X – ADH – P’ |
| 12 | 1 μg ALDH + Xbal + PstI | NEBuffer 2 | ‘X – ALDH – P’ |
| 13 | 3 μg pSB1T3 + EcoRI + PstI | NEBuffer 2 | ‘E – pSB1T3(lin) – P’ |
Emulsifier
Since last weeks attempt to construct the inducible protomer R0011 with RBS B0032 failed, we decided to take a different approach. Instead of doing the standard 2 parts assembly, we amplify the promoter by PCR digest it and ligate in the plasmid that contains the RBS which has only be cut open at the left side. By doing so we do not risk losing the super small DNA fragments like the RBS. The problem is that we cannot use antibiotics selection. So we need to determine that by Colony PCR and sequencing.
PCR Amplification
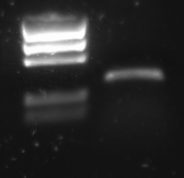
[http://partsregistry.org/Part:BBa_R0011 R0011] was amplified with the universal primers. The product was put on gel:
Lane description:
| # | Description | Amount |
| 1 | BioRad EZ Ladder | 5 ul |
| 2 | PCR Product | 10 ul + 2 ul LB |
The PCR band is about 300 bps long. R0011 itself is just 55 bp, but the flanking primer regions are about 100 bp each.
Digestion
The PCR product of promotor R0011 was cut with ... and the RBS containing plasmid has been cut open with ... at 37 C.
Ligation
The digestion products were ligated over night:
| # | Ligation reaction |
| 1 | 5 μL Cut plasmid with B0032 + 10 μL PCR Product of R0011 |
 "
"