|
DIY-GEM: a path towards low cost high throughput gene synthesis.
Synthetic biology research requires more cost effective approaches toward reagents and hardware accessibility. We are developing low-cost alternatives to existing hardware and enzymes in an attempt to expand participation in biological research and development. Our project expands the accessibility of Taq Polymerase by engineering it in a form compatible with BioBrick assembly. This allows use of the over-expressed enzyme from a crude bacterial extract in a PCR reaction at a fraction of the cost of highly purified commercial enzyme. In addition, we have developed inexpensive and easily assembled lab equipment such as a gel electrophoresis apparatus and a PCR thermal cycler. Enabling researchers to synthesize their own enzymes and having access to inexpensive tools will allow for increased participation among the DIY-bio community, stretch increasingly scarce educational funds, and allow rapid scale up of large scale gene synthesis projects."
Developing low-cost alternatives to existing enzymes: Taq polymerase Project Details
Thermus Aquaticus Polymerase I
PolI
J04639.1
Gene Sequence via BLAST at NCBI - http://www.ncbi.nlm.nih.gov/nuccore/155128
1 AAGCTCAGAT CTACCTGCCT GAGGGCGTCC GGTTCCAGCT GGCCCTTCCC
51 GAGGGGGAGA GGGAGGCGTT TCTAAAAGCC CTTCAGGACG CTACCCGGGG
101 GCGGGTGGTG GAAGGGTAAC ATGAGGGGGA TGCTGCCCCT CTTTGAGCCC
151 AAGGGCCGGG TCCTCCTGGT GGACGGCCAC CACCTGGCCT ACCGCACCTT
201 CCACGCCCTG AAGGGCCTCA CCACCAGCCG GGGGGAGCCG GTGCAGGCGG
251 TCTACGGCTT CGCCAAGAGC CTCCTCAAGG CCCTCAAGGA GGACGGGGAC
301 GCGGTGATCG TGGTCTTTGA CGCCAAGGCC CCCTCCTTCC GCCACGAGGC
351 CTACGGGGGG TACAAGGCGG GCCGGGCCCC CACGCCGGAG GACTTTCCCC
401 GGCAACTCGC CCTCATCAAG GAGCTGGTGG ACCTCCTGGG GCTGGCGCGC
451 CTCGAGGTCC CGGGCTACGA GGCGGACGAC GTCCTGGCCA GCCTGGCCAA
501 GAAGGCGGAA AAGGAGGGCT ACGAGGTCCG CATCCTCACC GCCGACAAAG
551 ACCTTTACCA GCTCCTTTCC GACCGCATCC ACGTCCTCCA CCCCGAGGGG
601 TACCTCATCA CCCCGGCCTG GCTTTGGGAA AAGTACGGCC TGAGGCCCGA
651 CCAGTGGGCC GACTACCGGG CCCTGACCGG GGACGAGTCC GACAACCTTC
701 CCGGGGTCAA GGGCATCGGG GAGAAGACGG CGAGGAAGCT TCTGGAGGAG
751 TGGGGGAGCC TGGAAGCCCT CCTCAAGAAC CTGGACCGGC TGAAGCCCGC
801 CATCCGGGAG AAGATCCTGG CCCACATGGA CGATCTGAAG CTCTCCTGGG
851 ACCTGGCCAA GGTGCGCACC GACCTGCCCC TGGAGGTGGA CTTCGCCAAA
901 AGGCGGGAGC CCGACCGGGA GAGGCTTAGG GCCTTTCTGG AGAGGCTTGA
951 GTTTGGCAGC CTCCTCCACG AGTTCGGCCT TCTGGAAAGC CCCAAGGCCC
1001 TGGAGGAGGC CCCCTGGCCC CCGCCGGAAG GGGCCTTCGT GGGCTTTGTG
1051 CTTTCCCGCA AGGAGCCCAT GTGGGCCGAT CTTCTGGCCC TGGCCGCCGC
1101 CAGGGGGGGC CGGGTCCACC GGGCCCCCGA GCCTTATAAA GCCCTCAGGG
1151 ACCTGAAGGA GGCGCGGGGG CTTCTCGCCA AAGACCTGAG CGTTCTGGCC
1201 CTGAGGGAAG GCCTTGGCCT CCCGCCCGGC GACGACCCCA TGCTCCTCGC
1251 CTACCTCCTG GACCCTTCCA ACACCACCCC CGAGGGGGTG GCCCGGCGCT
1301 ACGGCGGGGA GTGGACGGAG GAGGCGGGGG AGCGGGCCGC CCTTTCCGAG
1351 AGGCTCTTCG CCAACCTGTG GGGGAGGCTT GAGGGGGAGG AGAGGCTCCT
1401 TTGGCTTTAC CGGGAGGTGG AGAGGCCCCT TTCCGCTGTC CTGGCCCACA
1451 TGGAGGCCAC GGGGGTGCGC CTGGACGTGG CCTATCTCAG GGCCTTGTCC
1501 CTGGAGGTGG CCGAGGAGAT CGCCCGCCTC GAGGCCGAGG TCTTCCGCCT
1551 GGCCGGCCAC CCCTTCAACC TCAACTCCCG GGACCAGCTG GAAAGGGTCC
1601 TCTTTGACGA GCTAGGGCTT CCCGCCATCG GCAAGACGGA GAAGACCGGC
1651 AAGCGCTCCA CCAGCGCCGC CGTCCTGGAG GCCCTCCGCG AGGCCCACCC
1701 CATCGTGGAG AAGATCCTGC AGTACCGGGA GCTCACCAAG CTGAAGAGCA
1751 CCTACATTGA CCCCTTGCCG GACCTCATCC ACCCCAGGAC GGGCCGCCTC
1801 CACACCCGCT TCAACCAGAC GGCCACGGCC ACGGGCAGGC TAAGTAGCTC
1851 CGATCCCAAC CTCCAGAACA TCCCCGTCCG CACCCCGCTT GGGCAGAGGA
1901 TCCGCCGGGC CTTCATCGCC GAGGAGGGGT GGCTATTGGT GGCCCTGGAC
1951 TATAGCCAGA TAGAGCTCAG GGTGCTGGCC CACCTCTCCG GCGACGAGAA
2001 CCTGATCCGG GTCTTCCAGG AGGGGCGGGA CATCCACACG GAGACCGCCA
2051 GCTGGATGTT CGGCGTCCCC CGGGAGGCCG TGGACCCCCT GATGCGCCGG
2101 GCGGCCAAGA CCATCAACTT CGGGGTCCTC TACGGCATGT CGGCCCACCG
2151 CCTCTCCCAG GAGCTAGCCA TCCCTTACGA GGAGGCCCAG GCCTTCATTG
2201 AGCGCTACTT TCAGAGCTTC CCCAAGGTGC GGGCCTGGAT TGAGAAGACC
2251 CTGGAGGAGG GCAGGAGGCG GGGGTACGTG GAGACCCTCT TCGGCCGCCG
2301 CCGCTACGTG CCAGACCTAG AGGCCCGGGT GAAGAGCGTG CGGGAGGCGG
2351 CCGAGCGCAT GGCCTTCAAC ATGCCCGTCC AGGGCACCGC CGCCGACCTC
2401 ATGAAGCTGG CTATGGTGAA GCTCTTCCCC AGGCTGGAGG AAATGGGGGC
2451 CAGGATGCTC CTTCAGGTCC ACGACGAGCT GGTCCTCGAG GCCCCAAAAG
2501 AGAGGGCGGA GGCCGTGGCC CGGCTGGCCA AGGAGGTCAT GGAGGGGGTG
2551 TATCCCCTGG CCGTGCCCCT GGAGGTGGAG GTGGGGATAG GGGAGGACTG
2601 GCTCTCCGCC AAGGAGTGAT ACCACC
We took the above sequence from the provided link at BLAST and exported the SEQ into Plasma DNA. Plasma DNA is freeware from University of Helsinki which provides quick analysis of plasmid sequence information. http://research.med.helsinki.fi/plasmadna/
When we cut and paste this dna sequence into plasmadna and look at the output window, we are given a visual output of various coding information. Such as restriction sites found within the code. To consider a construct viable for a BbPart we'll need to make certain that the standard restriction enzymes used with the system won't sheer the dna making it incomplete code. Searching for EcoRI, Xbe1, Sbe1, Pst1 sites will show whether the code is viable in an untampered state.
Problem: PstI restriction site - Found @ 1717
CTGCAG-PstI restriction site
GACGTC-Complement
Solution - Site-specific Mutagenesis by Overlap Extension (see Sambrook, Joseph; Russell, David W. ; Molecular Cloning: A Laboratory Manual, 3rd Edition - http://www.cshlpress.com/default.tpl?cart=1279686078181232350&fromlink=T&linkaction=full&linksortby=oop_title&--eqSKUdatarq=21)
We then used the Gene Designer 2.0 freeware from DNA2.0 (https://www.dna20.com/genedesigner2/) - to analyze the Open Reading Frames. It shows us the Amino Acid codons that were being coded within that PstI Restrictions site. We find that the first three are coding for Leucine with CTG and can be changed at one point to CTT and still maintain Leucine's amino acid. The hope is that this will maintain functional integrity in the manufactured enzyme.
Primer Design
We designed two primers (11-14 Bp around chosen mutation) with changed Amino Acid Bp's Targeting initial Leucine at G of CTG to CTT. Point mutation Original G in CTG of Leucine. Change of one base to CTT maintains Leucine integrity.
GTGGAGAAGATCCT(T)CAGTACCGGCGG
CACCTCTTCTAGGA(A)GTCATGGCCGCC
While we're designing primers, besides the point mutation, we'll take the opportunity to design and order the primers for the Bb Suffix and Prefix. We'll follow the examples laid out in the Registry of Standard Parts under Promoter Construction for designing the oligos needed to make a part. (http://partsregistry.org/Help:Promoters/Construction)
Important considerations are Melting Point and percentage CG complements. Other considerations are dimerizations, that might cause primers to hairpin. We analyzed these primers using the OligoAnalyzer at IDT. When analyzing PolI Complements only were used for sequence inquiry, not the Bb Suffix/Prefixes. (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/)
PolI Coli Primers For Overlap Extension PCR
PCR Reaction 1
Bb Prefix + PolI (Fwd Complement) : (Forward complement will begin coding at 121 according to BLAST CDS information.)
GTTTCTTCGAATTCGCGGCCGCTTCTAGAG-ATGCTGCCCCTCTTTGAGCC
60.5 c ; 56.5 % GC Concetration
TAQ Rm
CTCCCGGTACTGAAGGATCTTCTCCAC
61.5 c ; 55.6 % GC Concentration
PCR Reaction - 2
TAQ Fm
GTGGAGAAGATCCTTCAGTACCGGGAG
61.5 c; 55.6 % GC
Bb Suffix + PolI (Reverse Complement) : (Reverse complement will end coding at 2619 according to Blast CDS information.
GTTTCTTCCTGCAGCGGCCGCTACTAGTA-TCACTCCTTGGCGGAGAGCC
61.8 c; 65 % GC
PCR Reaction - 3
Bb Prefix & Suffix Primers
Resuspend in 100 uL of H2O
Run PCR w 1/100 dilutions for PCR (5-10 uL per PCR reaction)
NEXT
- Create Full Bb Prmr w Plasmid combining new part using
<partinfo>R0010</partinfo> - Promoter (LacI)
<partinfo>B0034</partinfo> - Strong RBS
NEW PART - PolI Bb Format
<partinfo>B0015</partinfo> - Double Terminator
Psb1_?_3 - Plasmid of Interest with Chosen Resistance : http://partsregistry.org/Plasmid_backbones
<partinfo>R0010</partinfo> + <partinfo>B0034</partinfo> = New part LacI Promoter + Strong RBS
Cut <partinfo>R0010</partinfo> w/EcoRI & SpeI
Cut <partinfo>B0034</partinfo> w/XbeI & PstI
Combine in Chloramphenecol Resistant Plasmid (cut w/EcoRI & PstI) - Because
---
New Part + <partinfo>B0015</partinfo> = New Part
Cut New Part w/EcoRI & SpeI
Cut <partinfo>B0015</partinfo> w/XbeI & PstI
Combine in Chloramphenecol Resistant Plasmid (cut w/EcoRI & PstI)
Cut 1st Combined Part w/EcoRI & SpeI
Cut 2nd Combined Part w/XbeI & PstI
Combine in Ampecillan/Kanamyacin Resistan Plasmid (cut w/EcoRI & PstI)
Voila!!! Brand New Taq Polymerase Bb Part.
Developing low-cost alternatives to existing hardware: Project Details and Results
An unfortunate fact of reality is that precision lab equipment is very costly. Even simple devices such as an Electrophoresis or PCR have significant cost. To ameliorate this a portion of our project will involve designing biological tools that are easy to build and are economical.
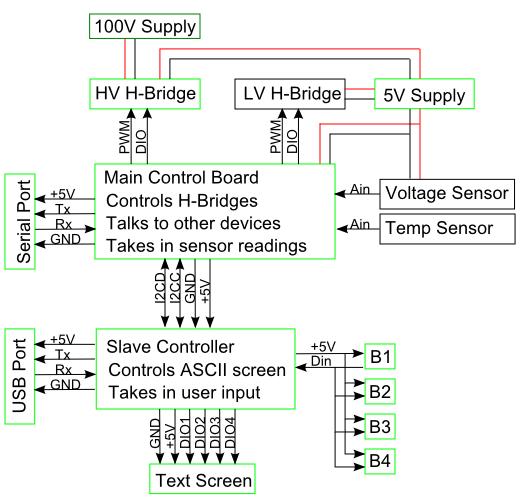
Our design incorporates two devices, a PCR and an Electrophoresis. Both are controlled by the same control electronics and power supply. A basic overview of the design can be seen in the diagram above. This design allows precise control from a computer or manual control from the control panel on the control electronics. Additionally multiple Electrophoresis devices can be controlled simultaneously in parallel and any power supply suitable can be used to power the devices.

With regards to equipment, we have successfully constructed a very low-cost Gel Electrophoresis device and are currently working on the control software and control electronics. Additionally, we are working on getting a low-cost PCR thermocycler up and running as well.
Instructions and Design files for building an Electrophoresis device
|
 "
"


