Team:USTC Software/detail
From 2010.igem.org
(Difference between revisions)
(→Details) |
|||
| Line 1: | Line 1: | ||
{{Team:USTC_Software/Header}} | {{Team:USTC_Software/Header}} | ||
| - | =Details= | + | =Modeling Details of Repressilator= |
| - | + | <br/> | |
| + | Key points of this modeling are: | ||
| + | * LacI protein tends to form LacI dimer, which tends to form LacI tetramer further. | ||
| + | * Only LacI tetramer binds with pLacI gene and thus repressing expression of its downstream genes. | ||
| + | * TetR protein only forms dimer. | ||
| + | * Only tetR dimer binds with pTet gene and thus repressing expression of its downstream genes. | ||
| + | * One IPTG molecule binds with one LacI tetramer to form complex IPTG:LacI4. | ||
| + | * cIlam protein only forms dimer. | ||
| + | * Only cIlam dimer binds with pcIlam gene and thus repressing expression of its downstream genes. | ||
| + | <br/> | ||
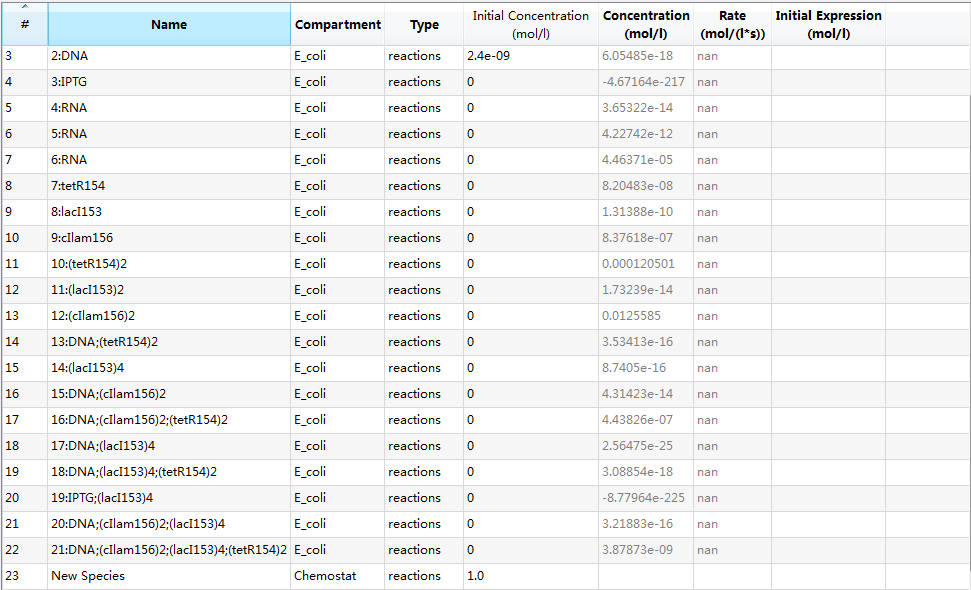
An overview of species list is provided: | An overview of species list is provided: | ||
| - | + | <br/> | |
[[Image:USTCS_graph1.JPG|600px|thumb|center]] | [[Image:USTCS_graph1.JPG|600px|thumb|center]] | ||
| - | + | <br/> | |
where each species with its specific identifier has its own meaning: | where each species with its specific identifier has its own meaning: | ||
| - | + | <br/> | |
*0:E_coli: E.Coli cell; | *0:E_coli: E.Coli cell; | ||
*1:IPTG: IPTG in flask; | *1:IPTG: IPTG in flask; | ||
Revision as of 16:41, 27 October 2010
Modeling Details of Repressilator
Key points of this modeling are:
- LacI protein tends to form LacI dimer, which tends to form LacI tetramer further.
- Only LacI tetramer binds with pLacI gene and thus repressing expression of its downstream genes.
- TetR protein only forms dimer.
- Only tetR dimer binds with pTet gene and thus repressing expression of its downstream genes.
- One IPTG molecule binds with one LacI tetramer to form complex IPTG:LacI4.
- cIlam protein only forms dimer.
- Only cIlam dimer binds with pcIlam gene and thus repressing expression of its downstream genes.
An overview of species list is provided:
where each species with its specific identifier has its own meaning:
- 0:E_coli: E.Coli cell;
- 1:IPTG: IPTG in flask;
- 2:DNA: initial transformed plasmids;
- 3:IPTG: IPTG in E.Coli due to diffusion;
- 4:RNA: mRNA of tetR;
- 5:RNA: mRNA of lacI;
- 6:RNA: mRNA of cIlam;
- 7:tetR154: tetR protein;
- 8:lacI153: lacI protein;
- 9:cIlam156: lacI protein;
- 10:(tetR154)2: tetR dimer;
- 11:(lacI153)2: lacI dimer;
- 12:(cIlam156)2: cIlam dimer;
- 13:DNA;(tetR154)2: complex of tetR dimer binding to pTet gene of plasmid DNA;
- 14:(lacI153)4; lacI tetramer;
- 15:DNA;(cIlam156)2: complex of cIlam dimer binding to pCI gene of plasmid DNA;
- 16:DNA;(cIlam156)2;(tetR154)2: complex of plasmid of DNA with both pCI and pTet genes bound with cIlam dimer and tetR dimer;
- 17:DNA;(lacI153)4: complex of LacI tetramer binding to pLacI gene plasmid DNA;
- 18:DNA;(lacI153)4;(tetR154)2: complex of plasmid DNA with both pLacI and pTet genes bound with LacI tetramer and tetR dimer, respectively;
- 19:IPTG;(lacI153)4: complex of LacI tetramer binding to pLacI gene of plasmid DNA;
- 20:DNA;(cIlam156)2;(lacI153)4: complex of plasmid DNA with both pCI and placI genes bound with cIlam dimer and lacI tetramer, respectively;
- 21:DNA;(cIlam156)2;(lacI153)4;(tetR154)2: complex of plasmid DNA with both pCI, placI and pTet genes bound with cIlam dimer, lacI tetramer and tetR dimer, respectively;
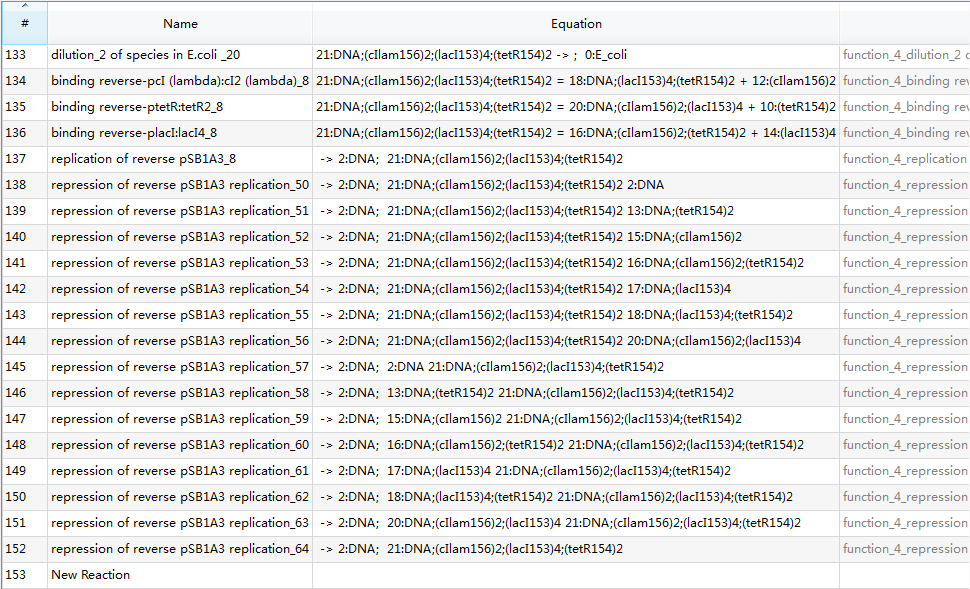
Since there are 153 reactions in total, we only provide a screenshot (we strongly recommend users to run our example to learn more details about automatic modeling):
 "
"