|
|
| (22 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{Team:USTC_Software/Header}} | | {{Team:USTC_Software/Header}} |
| | | | |
| - | ==One-minute introduction==
| + | = One-Minute Introduction = |
| - | ==Features==
| + | |
| - | ===Chain-Node Model===
| + | |
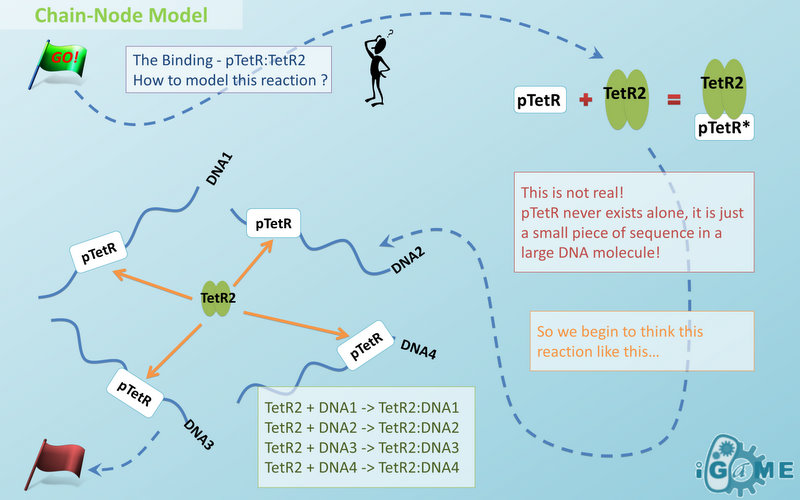
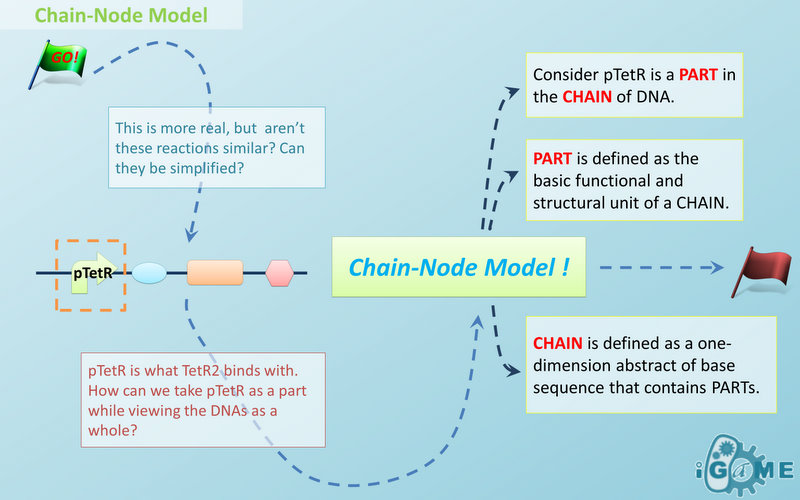
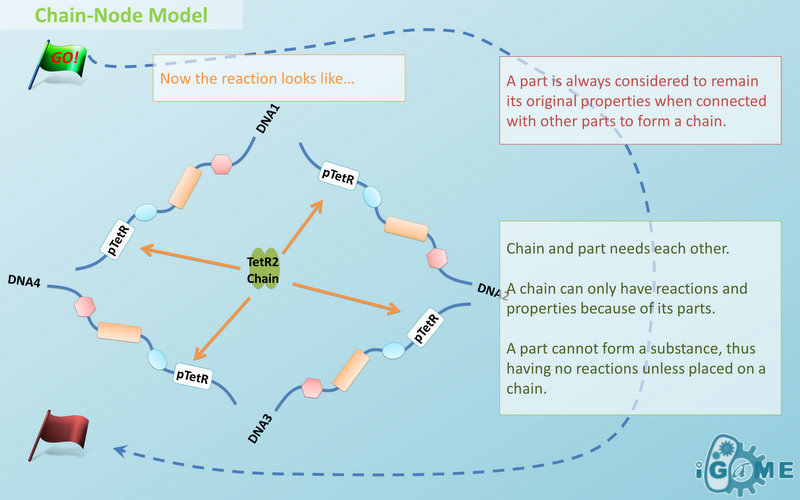
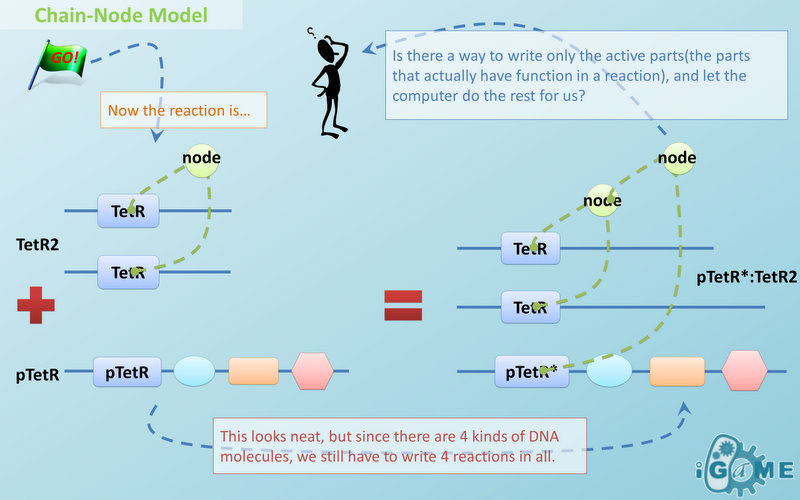
| - | In most manual modeling, complex is treated as a whole entity in reactions. Plenty of synthetic biology automatic modeling software treats complex the same way. However, there is a big difference between manual and automatic modeling, that is: Model constructors always know which detail of complex needs to be ignored or preserved, but software does not. So in most software, detailed structure has to be specified by modelers, otherwise will be ignored. In most cases, this requires plenty of manual operations and thus making the software not so automatic. Knowing this, we choose to preserve detailed structure in complex and present Chain-Node Model, which view complex as a construction of its parts throughout the modeling process. With chain and nodes, describing large biological complex with a great number of basic units and binding sites becomes possible. It also offers great flexibility in modeling. Detailed concepts are explained below.
| + | |
| | | | |
| - | ====Abstract concept of Chain====
| + | '''You are about to begin an exciting journey of the main ideas in MoDeL. For each picture, start from the green flag, follow the arrows, and finally you will reach the destination! Read on from the first step or click one of the three steps!''' |
| - | When it comes to biobrick assembly, DNA of plasmid is often considered as an abstract straight line where biobricks can be placed on and connected together. It actually acts as the basic frame of biobrick assembly. Inspired by this, we present the abstract concept of chain, which also acts as the basic frame of part assembly. Chain is an abstract straight line where different parts can be placed on and connected together. Part is the basic unit of chain, and chain can have as many parts as possible.
| + | <br /> |
| | + | <br /> |
| | + | <span style="font-size:6mm;">[https://2010.igem.org/Team:USTC_Software/MoDeL#Chain-Node_Model <font color="green">Chain-Node Model</font>] -> [https://2010.igem.org/Team:USTC_Software/MoDeL#Template_Modeling <font color="orange">Template Modeling</font>] -> [https://2010.igem.org/Team:USTC_Software/MoDeL#Automatic_Modeling_Database_Language <font color="blue">Automatic Modeling Database Language</font>]</span> |
| | + | <br /> |
| | + | [[Image:Ustcs Go.jpg|200px|center]] |
| | + | <br /> |
| | | | |
| - | To go beyond DNA, and to unify the data structure of Species, we extend the concept of chain to include all the parts we can describe, including RNA, protein, and even compartment, compound and substituent, etc. Parts of DNA, RNA and protein, are all originated from DNA, and they are considered as “Sequence”. Sequence can be connected to other parts of sequence on a chain. On the contrary, parts of compartment and compound are considered as “Non-sequence”. Parts of non-sequence cannot be connected to any other part on a chain, which means, they can only exist on a One-Part-Chain. Substituent is used as a substitution of any other part, so it can be either sequence or non-sequence.
| + | === <font color="green">Chain-Node Model</font> === |
| | + | [[Image:Ustcs WIKI widescreen 1.jpg|frame|center]] |
| | + | <br /> |
| | + | [[Image:Ustcs WIKI widescreen 2.jpg|frame|center]] |
| | + | <br /> |
| | + | [[Image:Ustcs WIKI widescreen 3.jpg|frame|center]] |
| | + | <br /> |
| | + | [[Image:Ustcs WIKI widescreen 4.jpg|frame|center]] |
| | + | <br /> |
| | + | [[Image:Ustcs WIKI widescreen 5.jpg|frame|center]] |
| | + | <br /> |
| | + | <p style="text-align: left;"><span style="font-size:4mm;">[[#top|Back Up]]</span></p> |
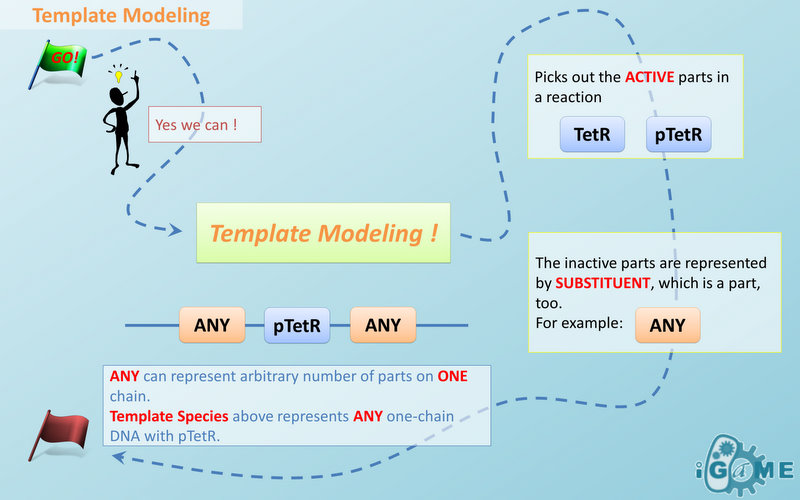
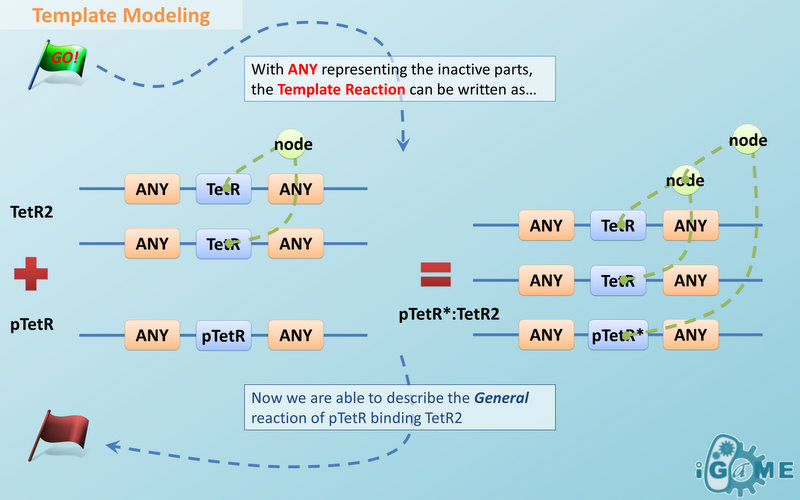
| | + | === <font color="orange">Template Modeling</font> === |
| | + | [[Image:Ustcs WIKI widescreen 6.jpg|frame|center]] |
| | + | <br /> |
| | + | [[Image:Ustcs WIKI widescreen 7.jpg|frame|center]] |
| | + | <br /> |
| | + | <p style="text-align: left;"><span style="font-size:4mm;">[[#top|Back Up]]</span></p> |
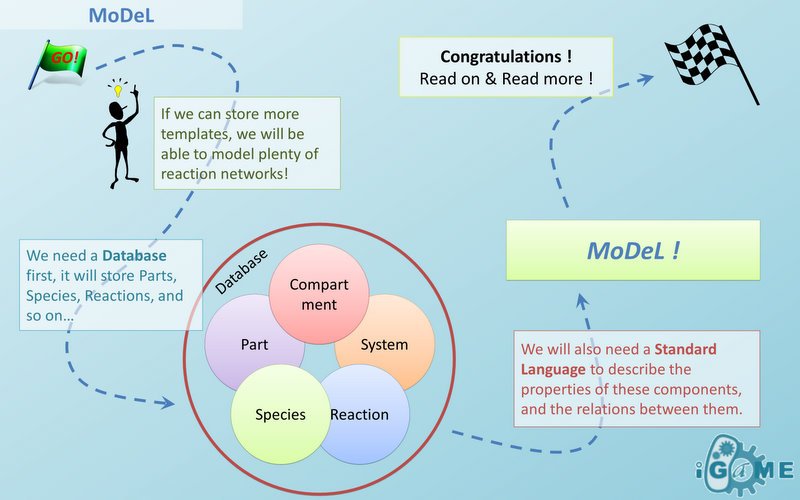
| | + | === <font color="blue">Automatic Modeling Database Language</font> === |
| | + | [[Image:Ustcs WIKI widescreen 8.jpg|frame|center]] |
| | + | <p style="text-align: left;"><span style="font-size:4mm;">[[#top|Back Up]]</span></p> |
| | + | <br /> |
| | + | [[Image:Ustcs Finish.jpg|200px|center]] |
| | | | |
| - | It is important here to mention that the Sequence/Non-sequence division is only a rule guide the modeling. It is not a constraint to Chain-Node model. Actually, we choose to maximally preserve the flexibility and leave all the freedom of constructing a species to users. Users are allowed to construct ANY species they can using Chain-Node Model. For example, one can create a strange species with compartment, compound, and DNA parts on the same chain, which makes little sense in biology. So, it is really up to the users to decide how to use Chain-Node Model in modeling.
| + | <span style="font-size:6mm;">Read More on MoDeL [https://2010.igem.org/Team:USTC_Software/model_features '''Here''']!</span> |
| - | | + | <span style="font-size:6mm;"> Read More on Our Project [https://2010.igem.org/Team:USTC_Software/Features '''Here''']!</span> |
| - | ====Abstract concept of Node==== | + | |
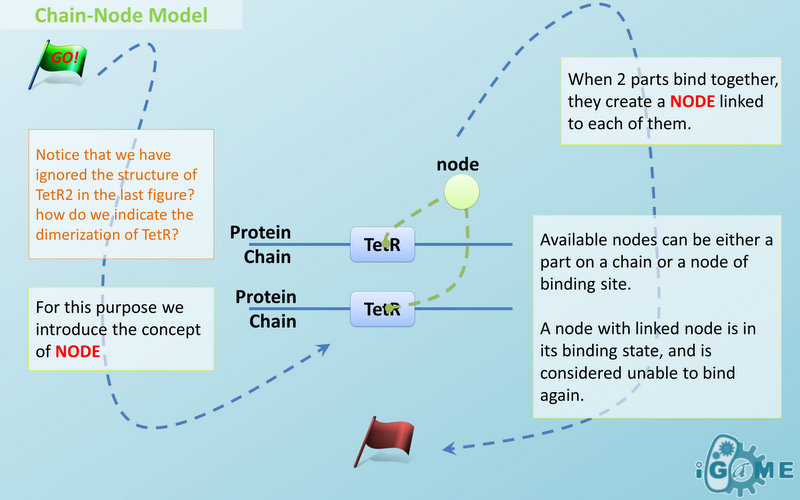
| - | Node is used to characterize binding structure in biological complex. Node is an abstract concept of binding site or site that can be bond. Links always exist between a binding site and the sites it binds together, indicating the relationship between them. We use trees to represent the binding structure in complex. The parent node represents the binding sites, and the child node represents the bond sites. For example the binding structure of TetR dimer is considered as a TetR2 node with two TetR nodes as its children. There is one exception though, the ROOT node. ROOT node is used as the top node in a tree. It can only have one child and no parent. The function of ROOT is to indicate that its child is the last binding site in this tree, and it binds to nothing any more.
| + | |
| - | | + | |
| - | In the reactions of promoter repression and activation, the promoter changes its properties after binding with repressor or inducer. It is not the original promoter any more. This is an example where binding changes the properties of its node. Since we do not store all the possible property alterations within one part, the solution is to replace the old part with a new part that stores the correct properties.
| + | |
| - | | + | |
| - | Available nodes can be both a part on a chain or a node of binding site, which leaves a lot of possibilities in constructing complicated binding structure. Again, there are no restrictions, but parts of compartments are not suggested to appear in a binding structure.
| + | |
| - | | + | |
| - | ====The basic assumption====
| + | |
| - | The basic assumption of Chain-Node Model is that a part always carries its properties wherever it is placed on a chain. This means no matter how complicated the complex is, you can always keep track of the location and properties of every single part. Letting complex inherits its parts’ properties, the assumption saves you from rewriting the structure and function of each new complex. However, we should address that the assumption is not suitable for all circumstances. For example, under this assumption, a fusion protein will carry the properties of the fused proteins, which is not true in many cases. A fusion protein may not remain its old properties and even have new properties that none of the original parts have. In this case, our suggestion is to define the fusion protein as a new part. So it is suggested that users choose different strategies of describing the same thing in Chain-Node Model base on different situations, instead of modeling everything in the same way.
| + | |
| - | | + | |
| - | ====Examples====
| + | |
| - | With great power there must come great responsibility. In order to use the powerful Chain-Node Model correctly, please have a look at a few examples we presents.
| + | |
| - | | + | |
| - | Example of species: E_coli_cell
| + | |
| - | | + | |
| - | Example of species: pLacI:lacI4 and its tree structure.
| + | |
| - | | + | |
| - | ===Template Reaction===
| + | |
| - | ===Auto-Modeling Database Language===
| + | |
| - | ==Language specifications==
| + | |
| - | ==History(optional)==
| + | |
 "
"