Team:Lethbridge/Notebook/Lab Work/September
From 2010.igem.org
JustinVigar (Talk | contribs) (→September 23, 2010) |
Liszabruder (Talk | contribs) (→AV) |
||
| (116 intermediate revisions not shown) | |||
| Line 57: | Line 57: | ||
</th> | </th> | ||
| - | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook"> | + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work"> |
<img src="https://static.igem.org/mediawiki/2010/7/73/UofLNotebookbutton.jpg" width="80"/> | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLNotebookbutton.jpg" width="80"/> | ||
</a> | </a> | ||
| Line 105: | Line 105: | ||
<body> | <body> | ||
<center> | <center> | ||
| - | <table border="0" width=" | + | <table border="0" width="28%" style="background-color:#000000"> |
<tr> | <tr> | ||
| + | <th> | ||
| + | <div class="miniBar"> | ||
| + | <div class="countdown"><object type="application/x-shockwave-flash" data="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=10&date_day=27&date_year=0&un=THE WIKI FREEZE&size=normal&mo=10&da=27&yr=2010" width="300" height="100"><param name="movie" value="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=10&date_day=27&date_year=0&un=THE WIKI FREEZE&size=normal&mo=10&da=27&yr=2010" /><param name="bgcolor" value="#000000" /></object><img src="http://www.oneplusyou.com/q/img/bb_badges/countdown.jpg" alt="" style="display: none;" height="1" width="1" /></div> | ||
| + | <div class="miniContainer"> | ||
| - | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook"> | + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work"> |
| - | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLNotebookbutton.jpg" width=" | + | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLNotebookbutton.jpg" width="80"/> |
</a> | </a> | ||
</th> | </th> | ||
| Line 116: | Line 120: | ||
<th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Protocols"> | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Protocols"> | ||
<img src="https://static.igem.org/mediawiki/2010/9/91/UofLprotocolsbutton.jpg" width="60"/> | <img src="https://static.igem.org/mediawiki/2010/9/91/UofLprotocolsbutton.jpg" width="60"/> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</a> | </a> | ||
</th> | </th> | ||
| Line 129: | Line 128: | ||
</th> | </th> | ||
| + | <th> | ||
| + | <div class="miniBar"> | ||
| + | <div class="countdown"><object type="application/x-shockwave-flash" data="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=11&date_day=05&date_year=0&un=THE IGEM JAMBOREE&size=normal&mo=11&da=05&yr=2010" width="300" height="100"><param name="movie" value="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=11&date_day=05&date_year=0&un=THE IGEM JAMBOREE&size=normal&mo=11&da=05&yr=2010" /><param name="bgcolor" value="#000000" /></object><img src="http://www.oneplusyou.com/q/img/bb_badges/countdown.jpg" alt="" style="display: none;" height="1" width="1" /></div> | ||
| + | <div class="miniContainer"> | ||
| + | </th> | ||
<tr> | <tr> | ||
</table> | </table> | ||
| Line 146: | Line 150: | ||
<body> | <body> | ||
<center> | <center> | ||
| - | <table border="0" width=" | + | <table border="0" width="50%" style="background-color:#000000"> |
<tr> | <tr> | ||
| Line 185: | Line 189: | ||
</th> | </th> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<tr> | <tr> | ||
| Line 198: | Line 199: | ||
<BLOCKQUOTE> | <BLOCKQUOTE> | ||
| + | =<font color="white">September= | ||
| + | ==<font color="white">September 2, 2010== | ||
| + | ===<font color="white">ADS=== | ||
| + | <b>Objective:</b> Assemble lumazine-dT into pSB1C3 using NEB BioBrick Assembly Kit.<br> | ||
| + | <b>Method:</b> Have the following parts: dT [28 ng/(µL)]; lumazine [78 ng/(µL)]; psB1C3 [14 ng/(µL)].<br> | ||
| + | *Require 500 ng DNA each part in a 50(µL) rxn therefore 17.9(µL) dT and 6.4(µL) Lumazine. Need 250 ng pSB1C3 or 17.9(µL) | ||
| - | + | Restriction Reactions - | |
| - | + | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b></td><td>Volume(µL)</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>36.1</td></tr> | ||
| + | <tr><td>NEBuffer 2 (10x)</td><td>5</td></tr> | ||
| + | <tr><td>Upstream Part (Lumazine)</td><td>26.4</td></tr> | ||
| + | <tr><td>EcoRI-HF</td><td>1</td></tr> | ||
| + | <tr><td>SpeI</td><td>1</td></tr> | ||
| + | <tr><td>100X BSA</td><td>0.5</td></tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b></td><td>Volume(µL)</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>24.6</td></tr> | ||
| + | <tr><td>NEBuffer 2 (10x)</td><td>5</td></tr> | ||
| + | <tr><td>Downstream Part (dT)</td><td>17.9</td></tr> | ||
| + | <tr><td>XbaI</td><td>1</td></tr> | ||
| + | <tr><td>PstI</td><td>1</td></tr> | ||
| + | <tr><td>100X BSA</td><td>0.5</td></tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b></td><td>Volume(µL)</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>3.35</td></tr> | ||
| + | <tr><td>NEBuffer 2 (10x)</td><td>2.5</td></tr> | ||
| + | <tr><td>Destination Plasmid (pSB1C3)</td><td>17.9</td></tr> | ||
| + | <tr><td>EcoRI-HF</td><td>.5</td></tr> | ||
| + | <tr><td>PstI</td><td>.5</td></tr> | ||
| + | <tr><td>100X BSA</td><td>0.25</td></tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | * 2(µL) was removed prior to adding enzymes for analysis on gel. | ||
| + | ** Split digestion reactions in half: dT - 2 @ 24(µL); Lum - 2 @ 24(µL); psB1C3 - 2 @ 11.5(µL) | ||
| + | *** Ran 10 minute and 60 minute reactions at 37<sup>o</sup>C. Followed by 20 minute heat shock at 80<sup>o</sup>C. | ||
| + | **** 2(µL) was removed from the heat killed reactions for analysis on gel. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | * Performed 4 combinations of ligation: 10 min Restriction + 10 min Ligation; 10 min Restriction + Overnight Ligation; 60 min Restriction + 10 min Ligation; 60 min Restriction + Overnight Ligation | ||
| + | |||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>1X(µL)</b><td><b>2X Master Mix(x2.5)(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>11<td>77.5 | ||
| + | <tr><td>T4 Ligase Buffer (10x)</td><td>2<td>5 | ||
| + | <tr><td>Upstream Part</td><td>2<td>5 | ||
| + | <tr><td>Downstream Part </td><td>2<td>5 | ||
| + | <tr><td>T4 DNA Ligase</td><td>1<td>2.5 | ||
| + | <tr><td>Plasmid Part</td><td>2<td>5 | ||
| + | </table> | ||
| + | |||
| + | Incubated 10 minute and overnight ligations at room temperature ( 25<sup>o</sup>C). Heat killed ligase at 80<sup>o</sup>C for 20 min. | ||
| + | * 2(µL) of each reaction was taken for analysis on gel. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | * Confirmed ligation with PCR analysis. Analyzed restriction, ligation and PCR on a 1.5% TAE agarose gel. | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x5.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>36.5<td>202.4 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>5<td>27.5 | ||
| + | <tr><td>dNTPs<td>2<td>11 | ||
| + | <tr><td>Forward VF2 Primer<td>2<td>11 | ||
| + | <tr><td>Reverse VR Primer<td>2<td>11 | ||
| + | <tr><td>Template DNA<td>2<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>1.1 | ||
| + | </table><br> | ||
| + | |||
| + | Added 48(µL) to each reaction. <br> | ||
| + | [[image:Lethbridge_100903AssemblyADS.jpg|200px]] | ||
| + | |||
| + | ---- | ||
| + | <b>Objective:</b> Characterize xylE in LB Media vs. M9 Minimal Media.<br> | ||
| + | <b>Method:</b> Three experiments with spectra. <br> | ||
| + | |||
| + | * Experiment 1 - Measure absorption spectra of 1:10 dilution of: 1. Cells suspended in m9 media +/- catechol. 2. Cells suspended in LB media +/- catechol. 3. m9 media from spun down cells +/- catechol. 4. LB media from spun down cells +/- catechol. 5. Sterile LB media +/- catechol. 6. Water +/- catechol. | ||
| + | |||
| + | * Experiment 2 - Catechol Breakdown. Follow absorbance of 2-hydroxymucanate semialdehyde at 375 nm. Measured all the above samples for 10 min at that wavelength. | ||
| + | |||
| + | ==<font color="white">September 14, 2010== | ||
| + | ===<font color="white">JF=== | ||
| + | <b>Objective:</b> Miniprep of RFP expression construct (J04450) via Qiaquick/Qiaprep spin column. Protocol followed as per kit instructions.<br> | ||
| + | |||
| + | ---- | ||
| + | ===<font color="white">JV=== | ||
| + | |||
| + | <b>Objective:</b> PCR the pSB1C3 plasmid from part J04450 miniprep.<br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>12.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2 | ||
| + | <tr><td>dNTPs<td>1 | ||
| + | <tr><td>SB-prep-26 Primer<td>1 | ||
| + | <tr><td>SB-prep-3P Primer<td>1 | ||
| + | <tr><td>Template DNA<td>2 | ||
| + | <tr><td>Pfu polymerase<td>0.2 | ||
| + | </table><br> | ||
| + | |||
| + | Used iGEM cycle PSB1C3 in thermocycler. | ||
| + | Ran PCR product on 1% TAE agarose gel. | ||
| + | |||
| + | ---- | ||
| + | ===<font color="white">KG=== | ||
| + | |||
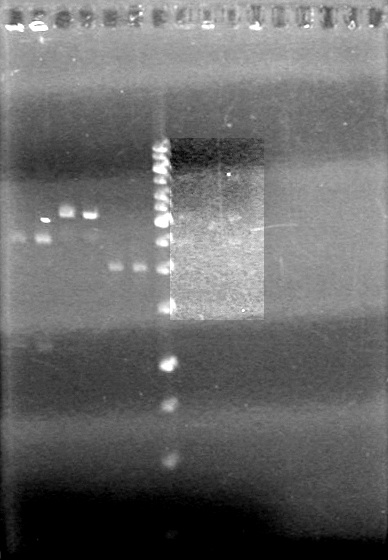
| + | <b>Objective:</b> PCR of xylE (mRBS-xylE K118021) with fusion standard so that an arginine tail can be attached.<br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x6)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>12.8<td>76.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>12 | ||
| + | <tr><td>dNTPs<td>1<td>6 | ||
| + | <tr><td>xylE Fusion Prefix Forward Primer<td>1<td>6 | ||
| + | <tr><td>xylE Fusion Suffix Reverse Primer<td>1<td>6 | ||
| + | <tr><td>Template DNA<td>2<td>12 | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>1.2 | ||
| + | </table><br> | ||
| + | |||
| + | PCR- Conditions: | ||
| + | 1. 95<sup>o</sup>C for 180 sec | ||
| + | 2. 95<sup>o</sup>C for 30 sec | ||
| + | 3. 56.6 +/- 5<sup>o</sup>C for 30 sec | ||
| + | 4. 72<sup>o</sup>C for 150 sec | ||
| + | 5. 72<sup>o</sup>C for 600 sec | ||
| + | (35 cycles) | ||
| + | |||
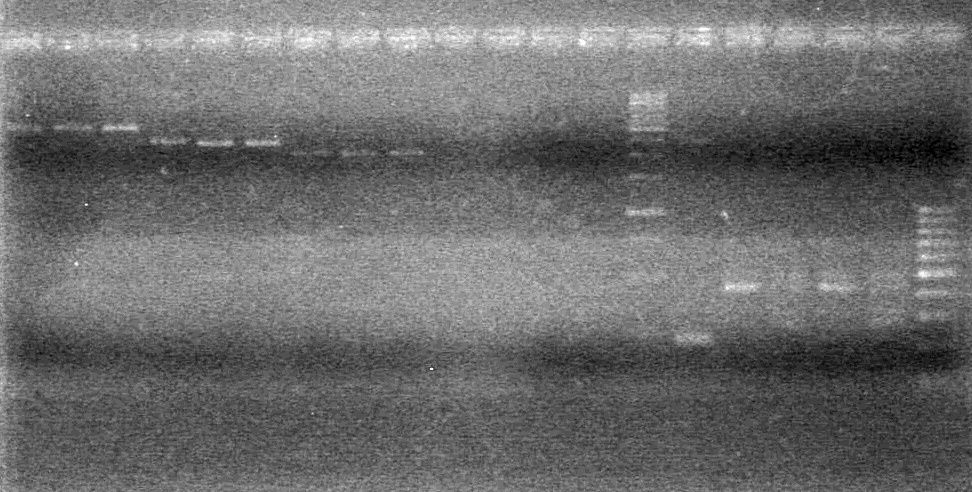
| + | Analyzed PCR on 2% agarose gel which ran at 100 V for 90 minutes.<br> | ||
| + | [[image:100915PCR AV KG JV.jpg|200px]] | ||
| + | |||
| + | ===<font color="white">AS=== | ||
| + | <b>Objective:</b> Assemble xylE-dT using 3AB assembly and 3AB assembly.<br> | ||
| + | |||
| + | Restriction Reactions - | ||
| + | |||
| + | 3AB Upstream Part (xylE) | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b></td><td>Volume(µL)</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>30.6</td></tr> | ||
| + | <tr><td>NEBuffer 2 (10x)</td><td>5</td></tr> | ||
| + | <tr><td>Plasmid DNA </td><td>11.9</td></tr> | ||
| + | <tr><td>EcoRI-HF</td><td>1</td></tr> | ||
| + | <tr><td>SpeI</td><td>1</td></tr> | ||
| + | <tr><td>100X BSA</td><td>0.5</td></tr> | ||
| + | </table> | ||
| + | |||
| + | 3AB Downstream Part (dT) | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b></td><td>Volume(µL)</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>24.6</td></tr> | ||
| + | <tr><td>NEBuffer 2 (10x)</td><td>5</td></tr> | ||
| + | <tr><td>Plasmid DNA</td><td>17.9</td></tr> | ||
| + | <tr><td>XbaI</td><td>1</td></tr> | ||
| + | <tr><td>PstI</td><td>1</td></tr> | ||
| + | <tr><td>100X BSA</td><td>0.5</td></tr> | ||
| + | </table> | ||
| + | |||
| + | 3AB Plasmid (pSB1C3) | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b></td><td>Volume(µL)</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td></td></tr> | ||
| + | <tr><td>NEBuffer 2 (10x)</td><td>5</td></tr> | ||
| + | <tr><td>Plasmid DNA</td><td>42.5</td></tr> | ||
| + | <tr><td>EcoRI-HF</td><td>1</td></tr> | ||
| + | <tr><td>PstI</td><td>1</td></tr> | ||
| + | <tr><td>100X BSA</td><td>0.5</td></tr> | ||
| + | </table> | ||
| + | |||
| + | *Ran 10 minute and 60 minute reactions at 37<sup>o</sup>C. Followed by 20 minute heat shock at 80<sup>o</sup>C. | ||
| + | |||
| + | Ligation Reaction - | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>1X(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>11 | ||
| + | <tr><td>T4 Ligase Buffer (10x)</td><td>2 | ||
| + | <tr><td>Upstream Part</td><td>2 | ||
| + | <tr><td>Downstream Part </td><td>2 | ||
| + | <tr><td>T4 DNA Ligase</td><td>1 | ||
| + | <tr><td>Plasmid Part</td><td>2 | ||
| + | </table> | ||
| + | |||
| + | Incubated 10 minute and overnight ligations at room temperature ( 25<sup>o</sup>C). Heat killed ligase at 80<sup>o</sup>C for 20 min. | ||
| + | |||
| + | Transformation - | ||
| + | |||
| + | A) Thawed 200(µL) Library Efficiency DH5alpha Competent Cells on ice. | ||
| + | B) Gently mixed cells and then aliquoted 100(µL) into chilled polypropylene tubes. | ||
| + | C) Added 1(µL) of ligation mix to cells. Added 5(µL) of pUC19 DNA to 100(µL) cells to determine efficiency. | ||
| + | D) Incubated cells on ice for 30 minutes. | ||
| + | E) Heat shocked cells for 45 seconds in a 42<sup>o</sup>C water bath. | ||
| + | F) Placed on ice for 2 minutes. | ||
| + | G) Added 0.9 mL of room temperature SOC medium. | ||
| + | H) Shook at 225 rpm for 1 hour. | ||
| + | I) Diluted control cells 1:100 with SOC medium. | ||
| + | J) Spread 100(µL) of this dilution on LB-Amp agar plates | ||
| + | K) Spread 50 and 250(µL) of experimental cells on LB-Cam agar plates. | ||
| + | L) Incubated overnight at 37<sup>o</sup>C | ||
| + | |||
| + | ==<font color="white">September 15, 2010== | ||
| + | ===<font color="white"> JV, ADS=== | ||
| + | <b>Objective:</b> Determine if dT ligated onto p-rbs-xylE using PCR analysis.<br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x4.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>11.8<td>53.1 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>9 | ||
| + | <tr><td>dNTPs<td>1<td>4.5 | ||
| + | <tr><td>Forward VF2 Primer<td>1<td>4.5 | ||
| + | <tr><td>Reverse VR Primer<td>1<td>4.5 | ||
| + | <tr><td>Template DNA<td>1<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td> | ||
| + | </table><br> | ||
| + | |||
| + | Used iGEM program 7 in thermocycler. | ||
| + | |||
| + | ---- | ||
| + | <b>Objective:</b> Determine if minipreps from Sept 15, 2010 worked. Analyze assembly intermediates. <br> | ||
| + | <b>Method:</b> Run 1% TAE agarose gels at 100 V for 90 minutes. <br> | ||
| + | |||
| + | [[image:100914ADSAssemblyPrep.jpg|100px]] | ||
| + | |||
| + | ==<font color="white">September 16, 2010== | ||
| + | ===<font color="white"> JV=== | ||
| + | <b>Objective:</b> Isolate RFP in pSB1C3 from DH5alpha cells.<br> | ||
| + | <b>Method:</b> Used Qiagen minipep protocol and spin column.<br> | ||
| + | <b>Results:</b> Gel of miniprepped RFP against previously prepared RFP showed a band of the right size.<br> | ||
| + | |||
| + | [[image:100916 JV RFP BW.jpg|50px]] | ||
| + | |||
| + | ==<font color="white">September 17, 2010== | ||
| + | ===<font color="white"> JV=== | ||
| + | <b>Objective:</b> Isolate plasmid DNA from DH5alpha cells: K249024; B0015; K118021/B0015<br> | ||
| + | <b>Method:</b> Used Qiagen minipep protocol and spin column. Ran a 1% agarose gel to view DNA<br> | ||
| + | |||
| + | GEL PICTURE! | ||
| + | ---- | ||
| + | ===<font color="white">AV=== | ||
| + | |||
| + | <b>Objective:</b> PCR out RFP from the complete construct (J04450) with a N-term fusion standard.<br> | ||
| + | |||
| + | Composition of each PCR tube: | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td>Volume(µL)</td></tr> | ||
| + | <tr><td>10x Pfu Buffer w/ MgSO<sub>4</sub></td><td>2</td></tr> | ||
| + | <tr><td>dNTP (10mM)</td><td>1</td></tr> | ||
| + | <tr><td>N-term Fus Prefix</td><td>1</td></tr> | ||
| + | <tr><td>Biobrick Suffix</td><td>1</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>12.8</td></tr> | ||
| + | <tr><td>Template DNA (J04450)</td><td>2</td></tr> | ||
| + | <tr><td>Pfu DNA Polymerase</td><td>0.2</td></tr> | ||
| + | </table> | ||
| + | |||
| + | PCR conditions: | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Step</b></td><td><b>Temperature (<sup>o</sup>C)</b></td><td><b>Time (mins)</b></td><td><b>Number of cycles</b></td></tr> | ||
| + | <tr><td>Initial Denaturation</td><td>95</td><td>2</td><td>1</td></tr> | ||
| + | <tr><td>Denaturation</td><td>95</td><td>0.5</td><td>25</td></tr> | ||
| + | <tr><td>Annealing</td><td>48.2, 52.4, 56.0 (gradient)</td><td>0.5</td><td>25</td></tr> | ||
| + | <tr><td>Extension</td><td>72</td><td>2</td><td>25</td></tr> | ||
| + | <tr><td>Final Extension</td><td>72</td><td>10</td><td>1</td></tr> | ||
| + | </table> | ||
| + | |||
| + | [[image:100915PCR AV KG JV.jpg|200px]] | ||
| + | |||
| + | Results: Amplification showing in gel, but fragment amplified was ~1000bp. RFP and dT should be ~800bp. Primers were checked and discovered the YFP/CFP primers were not compatible with the RFP. Concluded the amplification resulted from sloppy annealing of N-term fus prefix to the bio prefix resulting in the full construct being amplified (~1000bp) | ||
| + | ---- | ||
| + | |||
| + | ===<font color="white">KG=== | ||
| + | |||
| + | <b>Objective:</b> PCR xylE to attach fusion standard to the N-term and standard suffix to C-term.<br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x6)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>12.8<td>76.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>12 | ||
| + | <tr><td>dNTPs<td>1<td>6 | ||
| + | <tr><td>Fusion Forward Primer<td>1<td>6 | ||
| + | <tr><td>Standard Reverse Primer<td>1<td>6 | ||
| + | <tr><td>Template DNA<td>2<td>12 | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>1.2 | ||
| + | </table><br> | ||
| + | |||
| + | Fusion standard prefix has melting temperature of 66.5<sup>o</sup>C. Standard suffix has melting temperature of 70.5<sup>o</sup>C. | ||
| + | Annealing temperature of 61.5<sup>o</sup>C with 5 <sup>o</sup>C gradient. | ||
| + | PCR- Conditions: | ||
| + | 1. 95<sup>o</sup>C for 180 sec | ||
| + | 2. 95<sup>o</sup>C for 30 sec | ||
| + | 3. 61.5 +/- 5<sup>o</sup>C for 30 sec | ||
| + | 4. 72<sup>o</sup>C for 150 sec | ||
| + | 5. 72<sup>o</sup>C for 600 sec | ||
| + | (35 cycles) | ||
| + | |||
| + | ---- | ||
| + | <b>Objective:</b> PCR xylE to attach fusion standard to the C-term and standard suffix to N-term.<br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x6)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>12.8<td>76.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>12 | ||
| + | <tr><td>dNTPs<td>1<td>6 | ||
| + | <tr><td>Standard Forward Primer<td>1<td>6 | ||
| + | <tr><td>Fusion Reverse Primer<td>1<td>6 | ||
| + | <tr><td>Template DNA<td>2<td>12 | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>1.2 | ||
| + | </table><br> | ||
| + | |||
| + | Standard prefix has melting temperature of 68.7<sup>o</sup>C. Standard suffix has melting temperature of 61.6<sup>o</sup>C. | ||
| + | Annealing temperature of 56.6<sup>o</sup>C with 5 <sup>o</sup>C gradient. | ||
| + | |||
| + | PCR- Conditions: | ||
| + | 1. 95<sup>o</sup>C for 180 sec | ||
| + | 2. 95<sup>o</sup>C for 30 sec | ||
| + | 3. 56.6 +/- 5<sup>o</sup>C for 30 sec | ||
| + | 4. 72<sup>o</sup>C for 150 sec | ||
| + | 5. 72<sup>o</sup>C for 600 sec | ||
| + | (35 cycles) | ||
| + | |||
| + | ==<font color="white">September 18, 2010== | ||
| + | ===<font color="white">JS, DM=== | ||
| + | <b>Objective:</b> Overexpress mms6.<br> | ||
| + | Method: Followed overexpression protocol. One flask was induced with 250(µL) IPTG while another was induced with 50(µL). | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Time (minutes)</b></td><td><b>OD #1 (600λ)</b></td><td><b>OD #2 (600λ)</b></td></tr> | ||
| + | <tr><td>30</td><td>0.139</td><td>0.133</td></tr> | ||
| + | <tr><td>60</td><td>0.200</td><td>0.188</td></tr> | ||
| + | <tr><td>90</td><td>0.372</td><td>0.360</td></tr> | ||
| + | <tr><td>120</td><td>0.702</td><td>0.704</td></tr> | ||
| + | <tr><td>150(T<sub>0</sub>)</td><td>0.871</td><td>0.810</td></tr> | ||
| + | <tr><td>210(T<sub>1</sub>)</td><td>2.600</td><td>2.500</td></tr> | ||
| + | <tr><td>270(T<sub>2</sub>)</td><td>3.317</td><td>3.300</td></tr> | ||
| + | <tr><td>330(T<sub>3</sub>)</td><td>4.408</td><td>3.946</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | * Plated cells in presence of IPTG, Fe or IPTG and Fe on AMP(+) plates. Incubated at 37 <sup>o</sup>C. | ||
| + | ** Preparation of these samples was achieved by taking a 60(µL) cell sample and inoculating 5 mL LB media. These solutions were incubated at 37 <sup>o</sup>C for 30 min. To 1 mL of culture, the appropriate amount of Fe2+ [0.938(µL)], Fe3+[1.88(µL)] or IPTG [1(µL)] was added. | ||
| + | ---- | ||
| + | ===<font color="white">TF=== | ||
| + | |||
| + | <b>Objective:</b> Obtain preparations of B0015 (dT) and J04450 (RFP). | ||
| + | |||
| + | <b>Method:</b> Used [[Team:Lethbridge/Notebook/Protocols|Plasmid DNA Purification by Alkaline Lysis (Large Scale AKA Maxiprep]] protocol. | ||
| + | |||
| + | <b>Results:</b> Chloroform, DNA and isopropanol were mixed and therefore no products could be recovered. | ||
| + | ---- | ||
| + | |||
| + | |||
| + | |||
| + | ==<font color="white">September 20, 2010== | ||
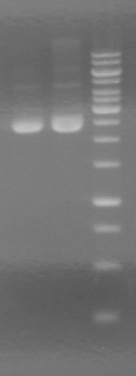
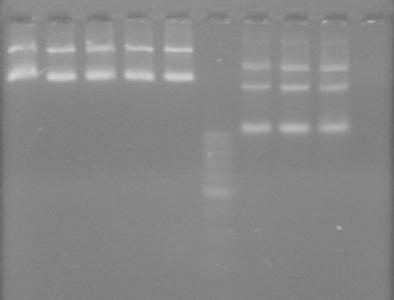
| + | ===<font color="white">ADS=== | ||
| + | <b>Objective:</b> Analyze PCR of mms6 (HB) and fluorescent proteins (AV).<br> | ||
| + | <b>Method:</b> Analyzed on 2% TAE agarose gel.<br> | ||
| + | Load order:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Volume<br>Sample (µL)</b></td><td><b>Volume Loading<br>Dye (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>HB1</td><td>10</td><td>2</td></tr> | ||
| + | <tr><td>2</td><td>HB2</td><td>10</td><td>2</td></tr> | ||
| + | <tr><td>3</td><td>HB3</td><td>10</td><td>2</td></tr> | ||
| + | <tr><td>4</td><td>HB4</td><td>10</td><td>2</td></tr> | ||
| + | <tr><td>5</td><td>HB5</td><td>10</td><td>2</td></tr> | ||
| + | <tr><td>6</td><td>100bp Ladder (Fermentas)</td><td>0.5</td><td>2(+10 H<sub>2</sub>0)</td></tr> | ||
| + | <tr><td>7</td><td>RFP1</td><td>10</td><td>2</td></tr> | ||
| + | <tr><td>8</td><td>RFP2</td><td>10</td><td>2</td></tr> | ||
| + | <tr><td>9</td><td>RFP3</td><td>10</td><td>2</td></tr></table><br> | ||
| + | <b>Results:</b> <br> | ||
| + | [[image:100920.jpg|100px]] | ||
| + | *mms6 was not amplified | ||
| + | *RFP was amplified | ||
| + | **Expected size is ~800bp | ||
| + | **Actual size is >1000bp | ||
| + | ***We believe that the suffix segment of the fusion primer annealed rather than the FP segment. This would cause the promoter (R0040) of J04450 to be amplified, adding ~200bp, giving a final size of >1000bp. | ||
| + | ---------------------------------------------------------------------------------------------------------------------- | ||
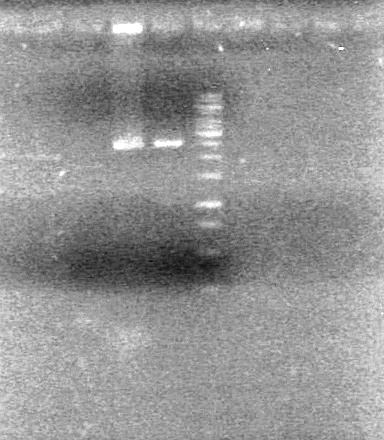
| + | <b>Objective:</b> Confirm assembly (3 antibiotic) of K118021 and B0015 from Sept. 14, 2010 via PCR.<br> | ||
| + | <b>Method:</b> Set up 50uL reaction mixture with 2uL of ligated DNA. Used VF2 and VR primers.<br> | ||
| + | Used PFU setting on Thermocycler<br> | ||
| + | <b>Results:</b> <br> | ||
| + | [[image:100915Assembly.jpg|100px]] | ||
| + | <b>Conclusion:</b> Assembly did not work as intended. | ||
| + | ---- | ||
| + | |||
| + | ===<font color="white">HB=== | ||
| + | |||
| + | <b>Objective:</b> PCR mms6 to attach fusion standard to the N-term and standard suffix to C-term.<br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x6)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>12.8<td>76.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>12 | ||
| + | <tr><td>dNTPs<td>1<td>6 | ||
| + | <tr><td>Fusion Forward Primer<td>1<td>6 | ||
| + | <tr><td>Standard Reverse Primer<td>1<td>6 | ||
| + | <tr><td>Template DNA<td>2<td>12 | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>1.2 | ||
| + | </table><br> | ||
| + | |||
| + | Prefix fusion sense primer has melting temperature of 56.1<sup>o</sup>C. Standard suffix primer has melting temperature of 61.3<sup>o</sup>C. | ||
| + | |||
| + | PCR- Conditions: | ||
| + | 1. 95<sup>o</sup>C for 180 sec | ||
| + | 2. 95<sup>o</sup>C for 30 sec | ||
| + | 3. 51.1<sup>o</sup>C for 30 sec | ||
| + | 4. 72<sup>o</sup>C for 150 sec | ||
| + | 5. 72<sup>o</sup>C for 600 sec | ||
| + | (35 cycles) | ||
| + | |||
| + | Program HBPREFU: | ||
| + | 1. 49.1<sup>o</sup>C 2. 50.2<sup>o</sup>C 3. 51.4<sup>o</sup>C 4. 52.6<sup>o</sup>C 5. 53.5<sup>o</sup>C | ||
| + | |||
| + | ==<font color="white">September 21, 2010== | ||
| + | ===<font color="white">ADS=== | ||
| + | <b>Objective:</b> Insert xylE (with N and C terminal fusion standards, obtained by PCR of K118021 by KG) into pSB1C3 plasmid for submission to registry.<br> | ||
| + | |||
| + | <b>Method:</b> | ||
| + | *Restriction of xylE PCR product and pSB1C3 (containing J04450 biobrick) via BioBrick Method using EcoRI-HF and PstI (Enzymes from NEB) | ||
| + | *Ligation of xylE PCR product and pSB1C3 via BioBrick Method using T4 DNA ligase (Enzyme from NEB) | ||
| + | *Transform into Library Efficiency Compentent DH5α Cells (Invitrogen) | ||
| + | |||
| + | <b>Results:</b> | ||
| + | *Obtained too numerous to count (TNTC) colonies. | ||
| + | **Obtained ~100 white colonies (indicating removal of RFP and insertion of new BioBrick) | ||
| + | |||
| + | <b>Note:</b><br> | ||
| + | Parent plasmid (from PCR) not digested, possibly K118021 moved into pSB1C3 backbone. | ||
| + | |||
| + | <b>Follow-up:</b><br> | ||
| + | Screen white colonies by addition of catechol to solution containing white cells. | ||
| + | ---- | ||
| + | ===<font color="white">TF, MC=== | ||
| + | |||
| + | <b>Objective:</b> Obtain a preparation of B0015 (dT). | ||
| + | |||
| + | <b>Method:</b> Used [[Team:Lethbridge/Notebook/Protocols|Plasmid DNA Purification by Alkaline Lysis (Large Scale AKA Maxiprep]] protocol. | ||
| + | ---- | ||
| + | ===<font color="white">AV=== | ||
| + | <b>Objective:</b> PCR amplify the signal sequences synthesized by Mr. Gene (K331007, K331008, K331009, K331012).<br> | ||
| + | |||
| + | Composition of each PCR tube: | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume(µL)</b></td></tr> | ||
| + | <tr><td>10x Pfu Buffer w/ MgSO<sub>4</sub></td><td>2</td></tr> | ||
| + | <tr><td>dNTP (10mM)</td><td>1</td></tr> | ||
| + | <tr><td>VF2</td><td>1</td></tr> | ||
| + | <tr><td>VR</td><td>1</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>12.8</td></tr> | ||
| + | <tr><td>Template DNA</td><td>2</td></tr> | ||
| + | <tr><td>Pfu DNA Polymerase</td><td>0.2</td></tr> | ||
| + | </table><br> | ||
| + | Used iGEM thermocycler setting: PFU.<br> | ||
| + | |||
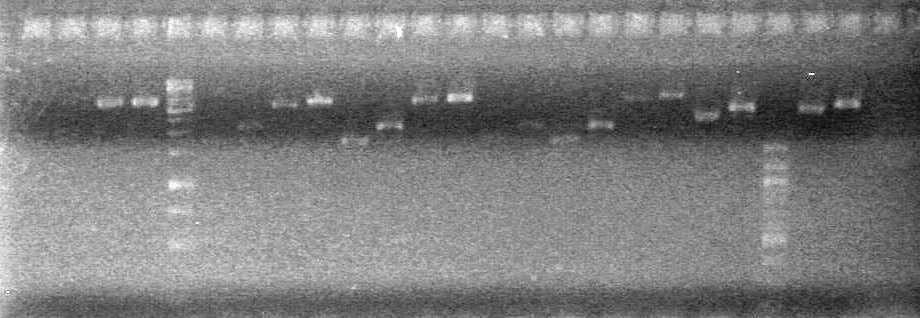
| + | ==<font color="white">September 21, 2010== | ||
| + | ===<font color="white">ADS=== | ||
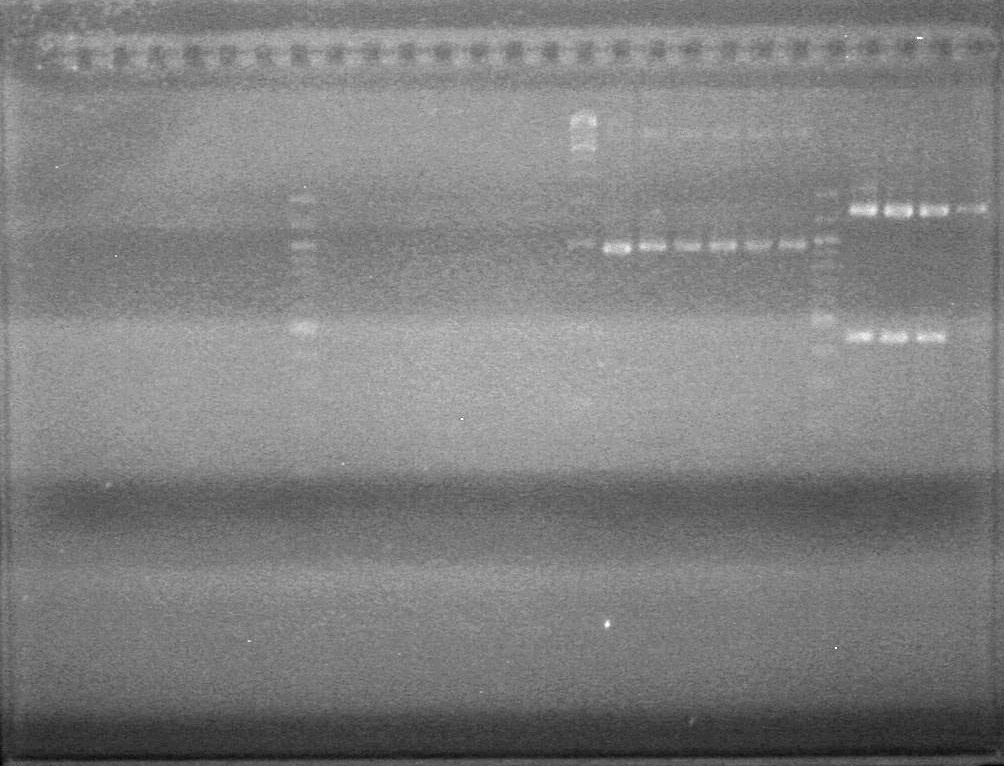
| + | <b>Objective:</b> Analyze PCR of K118021-B0015 and subsequent PCR (ADS) and PCR of <partinfo>K118021</partinfo> to add either N or C terminal fusion standard (KG).<br> | ||
| + | <b>Method:</b> Analyze on 2% TAE agarose gel.<br> | ||
| + | Load Order:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Volume<br>Sample (µL)</b></td><td><b>Volume Loading<br>Dye (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>xylE F-S 1</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>2</td><td>xylE F-S 2</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>3</td><td>xylE F-S 3</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>4</td><td>xylE F-S 4</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>5</td><td>xylE F-S 5</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>6</td><td>xylE F-S 6</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>7</td><td>100bp Ladder (NEB)</td><td>0.5</td><td>1(+5 H<sub>2</sub>0)</td></tr> | ||
| + | <tr><td>8</td><td>xylE S-F 1</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>9</td><td>xylE S-F 2</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>10</td><td>xylE S-F 3</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>11</td><td>xylE S-F 4</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>12</td><td>xylE S-F 5</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>13</td><td>xylE S-F 6</td><td>5</td><td>1</td></tr> | ||
| + | <tr><td>14</td><td>Empty</td><td></td><td></td></tr> | ||
| + | <tr><td>15</td><td>PCR of K118021-B0015 (ADS)</td><td>1</td><td>1</td></tr> | ||
| + | <tr><td>16</td><td>Empty</td><td></td><td></td></tr> | ||
| + | <tr><td>17</td><td>Empty</td><td></td><td></td></tr></table><br> | ||
| + | <b>Results:</b> <br> | ||
| + | [[image:100921.jpg|200px]] | ||
| + | *Both KG PCR (Standard-Fusion and Fusion-Standard) amplified | ||
| + | *ADS PCR Amplified, but no insert present. | ||
| + | |||
| + | ==<font color="white">September 23, 2010== | ||
| + | ===<font color="white">JV=== | ||
<b>Objective:</b> Characterized catechol degradation by xylE enzyme<br> | <b>Objective:</b> Characterized catechol degradation by xylE enzyme<br> | ||
| Line 210: | Line 698: | ||
*2) Take 1/10 dilution of cells | *2) Take 1/10 dilution of cells | ||
*3) Introduce 1µL of 0.05M catechol solution into the cell dilution. (Final concentration of 50µM;). | *3) Introduce 1µL of 0.05M catechol solution into the cell dilution. (Final concentration of 50µM;). | ||
| - | *4) Quench the reaction with 5A% w/v trichloroacetate at certain time points. (0,15sec, 30sec, 45sec, 60sec, 2min). | + | *4) Quench the reaction with 5A% w/v trichloroacetate at certain time points. (0,15sec, 30sec, 45sec, 60sec, 2min, 3min, 4min, 5min, 10min). |
*5) Spin down cells. | *5) Spin down cells. | ||
*6) Measure absorbance of supernatant.<br> | *6) Measure absorbance of supernatant.<br> | ||
| - | <b>Results:</b> | + | <b>Results: Cuvette used interfered with Spectra. </b> |
| + | |||
| + | ===<font color="white">ADS=== | ||
| + | <b><font size="+1">NOTE:</font></b> In all transformations, heat shock step was missed. HOWEVER, all transformations showed significant number of colony forming units.<br> | ||
| + | <b>Objective:</b> Move xylE (two biobrick; one with Fusion prefix, one with fusion suffix) into pSB1C3.<br> | ||
| + | <b>Method:</b> | ||
| + | *Restriction of xylE PCR product and pSB1C3 (containing J04450 biobrick) via BioBrick Method using EcoRI-HF and PstI (Enzymes from NEB) | ||
| + | *Ligation of xylE PCR product and pSB1C3 via BioBrick Method using T4 DNA ligase (Enzyme from NEB) | ||
| + | **Incubated 30 min at RT | ||
| + | *Transform into Subcloning Efficiency Compentent DH5α Cells (Invitrogen) | ||
| + | <b>Results:</b> TBD <br> | ||
| + | <b>Follow-up:</b> TBD<br> | ||
| + | ---------------------------------------------------------------------------------------------------------------------- | ||
| + | <b>Objective:</b> Create glycerol stocks of <partinfo>J04450</partinfo> in pSB1A3 and pSB1T3 for use in RFP-BioBrick Assembly.<br> | ||
| + | <b>Method:</b> Transform into Subcloning Efficiency Competent DH5α Cells (Invitrogen)<br> | ||
| + | Obtained all plasmid DNA from 2010 Kit Plate 1 | ||
| + | *J04450 in pSB1A3 - Well 1C | ||
| + | *J04450 in pSB1T3 - Well 7A | ||
| + | <b>Results:</b> Obtained TNTC colonies<br> | ||
| + | <b>Follow-up:</b> | ||
| + | *Grow overnight cultures | ||
| + | *Generate Glycerol Stocks | ||
| + | *Generate Plasmid DNA via Maxiprep | ||
| + | ---------------------------------------------------------------------------------------------------------------------- | ||
| + | <b>Objective:</b> Create glycerol stocks of received synthesized (Mr. Gene) signal peptides.<br> | ||
| + | <b>Method:</b> Transform into Subcloning Efficiency Competent DH5α Cells (Invitrogen) plasmid DNA containing the following BioBricks: | ||
| + | *1) <partinfo>K331007</partinfo> - β-lactamase Bla Signal Sequence | ||
| + | *2) <partinfo>K331008</partinfo> - Outer Membrane Protein ompA | ||
| + | *3) <partinfo>K331009</partinfo> - Heat Stable Toxin I | ||
| + | *4) <partinfo>K331012</partinfo> - Penicillin Binding Protein DacA | ||
| + | *All inserts in pMA-T vector (Standard Mr. Gene vector) | ||
| + | <b>Results:</b> Obtained TNTC Cells <br> | ||
| + | <b>Follow-up:</b> | ||
| + | *1) Grow overnight cultures | ||
| + | *2) Purify pDNA | ||
| + | *3) Move into pSB1C3 plasmid | ||
| + | *4) Verify sequence | ||
| + | *5) Submit to registry for sequencing | ||
| + | ---- | ||
| + | ===<font color="white">TF, MC=== | ||
| + | |||
| + | <b>Objective:</b> Obtain a preparation of J04450 (RFP).<br> | ||
| + | |||
| + | <b>ethod:</b> Used [[Team:Lethbridge/Notebook/Protocols|Plasmid DNA Purification by Alkaline Lysis (Large Scale AKA Maxiprep]] protocol. | ||
| + | |||
| + | ==<font color="white">September 24, 2010== | ||
| + | ===<font color="white">ADS=== | ||
| + | <b>Objective:</b> Generate plasmid DNA of <partinfo>E1010</partinfo> for downstream PCR<br> | ||
| + | <b>Method:</b> Transform plasmid DNA into Subcloning Efficiency Competent DH5α Cells (Invitrogen)<br> | ||
| + | DNA obtained from 2010 Kit Plate 1 Well 18F (E1010 in pSB2K3)<br> | ||
| + | <b>Results:</b> Obtained <font color="red"> TBD</font> colonies<br> | ||
| + | <b>Follow-up:</b> | ||
| + | *Grow overnight cultures (Generate glycerol stocks) | ||
| + | *Purify plasmid DNA (Generate pDNA stocks) | ||
| + | *PCR to add terminal fusion standards | ||
| + | ---------------------------------------------------------------------------------------------------------------------- | ||
| + | <b>Objective:</b> Create glycerol stocks of J04450 in pSB1K3 for use in RFP-BioBrick Assembly.<br> | ||
| + | <b>Method:</b> Transform into Subcloning Efficiency Competent DH5α Cells (Invitrogen)<br> | ||
| + | Obtained plasmid DNA from 2010 Kit Plate 1 well 5A (J04450 in pSB1K3)<br> | ||
| + | <b>Results:</b> Obtained TNTC colonies<br> | ||
| + | <b>Follow-up:</b> | ||
| + | *Grow overnight cultures | ||
| + | *Generate Glycerol Stocks | ||
| + | *Generate Plasmid DNA via Maxiprep | ||
| + | |||
| + | ==<font color="white">September 25, 2010== | ||
| + | ===<font color="white">JV=== | ||
| + | <b>Objective:</b> Extract Plasmid DNA from DH5α cells.<br> | ||
| + | |||
| + | <b>Method:</b>Qiagen spin column protocol.<br> | ||
| + | |||
| + | *<partinfo>K331007</partinfo> (in pMA-T vector) | ||
| + | *<partinfo>K331008</partinfo> (in pMA-T vector) | ||
| + | *<partinfo>K331009</partinfo> (in pMA-T vector) | ||
| + | *<partinfo>K331012</partinfo> (in pMA-T vector)<br> | ||
| + | |||
| + | Cells containing plasmids were put into glycerol stocks and put into HJ's -80<sup>o</sup>C.<br> | ||
| + | |||
| + | ===<font color="white">ADS=== | ||
| + | <b>Objective:</b> Move plasmid DNA made by JV (above, Sept 25, 2010) into pSB1C3 for submission to the Standard Registry of Parts.<br> | ||
| + | <b>Method:</b> | ||
| + | *1) Digest at 37<sup>o</sup> for 10 min with PstI and EcoRI: | ||
| + | **pSB1C3 containing <partinfo>J04450</partinfo> | ||
| + | **pMA-T with <partinfo>K331007</partinfo>, <partinfo>K331008</partinfo>, <partinfo>K331009</partinfo>, and <partinfo>K331012</partinfo>. | ||
| + | *2) Heat Kill at 80<sup>o</sup>C for 20 min | ||
| + | *3) Mix 2µL of pSB1C3 with each biobrick, ligate with T4 DNA ligase for 10 min | ||
| + | *4) Transform into subcloning efficiency DH5α cells, plate on ampicillin plates.<br> | ||
| + | <b>Results:</b> Obtained <font color="red">RESULT!!!</font color> positive colonies<br> | ||
| + | ---------------------------------------------------------------------------------------------------------------------- | ||
| + | <b>Objective:</b> Assemble KG's C-terminal fusion xylE (<partinfo>K331021</partinfo>) (<font color="red">UNCONFIRMED</font>) and C-terminal Arginine + dT (<partinfo>S04261</partinfo>) to create <partinfo>K331011</partinfo>. <br> | ||
| + | <b>Method:</b> | ||
| + | *1) Digest at 37<sup>o</sup> for 10 min: | ||
| + | **pSB1C3 containing <partinfo>J04450</partinfo> with PstI and EcoRI | ||
| + | **<partinfo>K331021</partinfo> with EcoRI, SpeI | ||
| + | **<partinfo>S04261</partinfo> with XbaI and PstI | ||
| + | *2) Heat Kill at 80<sup>o</sup>C for 20 min | ||
| + | *3) Mix 2µL of pSB1C3 with each biobrick, ligate with T4 DNA ligase for 10 min | ||
| + | *4) Transform into subcloning efficiency DH5α cells, plate on ampicillin plates.<br> | ||
| + | <b>Results:</b> Obtained <font color="red">RESULT!!!</font color> positive colonies<br> | ||
| + | |||
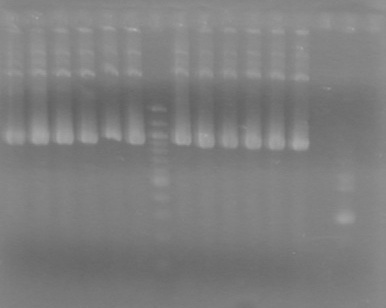
| + | ==<font color="white">September 26, 2010== | ||
| + | ===<font color="white">ADS=== | ||
| + | <b>Objective:</b> Purify plasmid DNA from 5mL cultures grown from transformations of KG's ligated PCRs containing (<font color="red">UNCONFIRMED</font>) <partinfo>K331020</partinfo> and <partinfo>K331021</partinfo>.<br> | ||
| + | <b>Method:</b> Used Qiagen spin column method with BioBasic EZ-10 spin columns.<br> | ||
| + | ------------------------------------------------------------------------------------------------------------------------ | ||
| + | <b>Objective:</b> Assemble the following:<br> | ||
| + | *<partinfo>K331020</partinfo> + <partinfo>B0015</partinfo> to obtain <font color="red"> To be named</font> biobrick | ||
| + | *<partinfo>K331021</partinfo> + <partinfo>S04261</partinfo> to obtain <font color="red"> To be named</font> biobrick | ||
| + | <b>Method:</b><br> | ||
| + | *Digest <partinfo>K331020</partinfo> and <partinfo>K331021</partinfo> with EcoRI and SpeI | ||
| + | *Digest <partinfo>B0015</partinfo> and <partinfo>S04261</partinfo> with XbaI and PstI | ||
| + | *Digest <partinfo>J04450</partinfo> in pSB1C3 with EcoRI and PstI | ||
| + | Incubate each at 37<sup>o</sup>C for 10 min<br> | ||
| + | Heat kill at 80<sup>o</sup>C for 20 min <br> | ||
| + | Ligate with T4 Ligase for 10 min at room temperature<br> | ||
| + | Transform into DH5&alpha subcloning efficiency cells<br> | ||
| + | <b>Results:</b> Obtained <font color="red">RESULTS!!!!</font color> positive colonies.<br> | ||
| + | Also analyzed minipreps on 1.5% TAE Agarose Gel<br> | ||
| + | [[image:100926Minipreps.jpg|200px]] | ||
| + | |||
| + | ==<font color="white">September 29, 2010== | ||
| + | ===<font color="white">ADS=== | ||
| + | Received synthesized parts from Mr. GENE (all in pMA vectors)<br> | ||
| + | *<partinfo>K331022</partinfo> | ||
| + | *<partinfo>K331023</partinfo> | ||
| + | *<partinfo>K331024</partinfo> | ||
| + | *<partinfo>K331025</partinfo> | ||
| + | Each tube contained 5µg pDNA; reconstitute in 50µL of TE buffer to get 100ng/µL concentration<br> | ||
| + | Transform 1µL into DH5α subcloning efficiency cells<br> | ||
| + | Obtained TNTC colonies<br> | ||
| + | Started overnight 5mL cultures for miniprep and insertion into pSB1C3 vector. | ||
| + | |||
| + | ---- | ||
| + | ===<font color="white">AV=== | ||
| + | |||
| + | <b>Objective:</b> PCR the N-term fusion tag onto RFP (E1010) using sloppy annealing of the YFP/CFP fusion primers. <br><br> | ||
| + | |||
| + | Composition of each PCR tube: | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td>Volume(µL)</td></tr> | ||
| + | <tr><td>10x Pfu Buffer w/ MgSO<sub>4</sub></td><td>2</td></tr> | ||
| + | <tr><td>dNTP (10mM)</td><td>1</td></tr> | ||
| + | <tr><td>N-term Fus Prefix</td><td>1</td></tr> | ||
| + | <tr><td>Biobrick Suffix</td><td>1</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>12.8</td></tr> | ||
| + | <tr><td>Template DNA (J04450)</td><td>2</td></tr> | ||
| + | <tr><td>Pfu DNA Polymerase</td><td>0.2</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | PCR conditions: | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Step</b></td><td><b>Temperature (<sup>o</sup>C)</b></td><td><b>Time (mins)</b></td><td><b>Number of cycles</b></td></tr> | ||
| + | <tr><td>Initial Denaturation</td><td>95</td><td>2</td><td>1</td></tr> | ||
| + | <tr><td>Denaturation</td><td>95</td><td>0.5</td><td>25</td></tr> | ||
| + | <tr><td>Annealing</td><td>47.6, 50.5, 53.8 (gradient)</td><td>0.5</td><td>25</td></tr> | ||
| + | <tr><td>Extension</td><td>72</td><td>2</td><td>25</td></tr> | ||
| + | <tr><td>Final Extension</td><td>72</td><td>10</td><td>1</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | Results: Gel has not been run. Have decided to order RFP specific primers for next year.<br><br> | ||
| + | <br> | ||
 "
"