Team:Lethbridge/Notebook/Lab Work/July
From 2010.igem.org
(→July 8/2010 - Evening) |
Liszabruder (Talk | contribs) |
||
| (87 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | <div style="background-color:#000000; color:white"> | |
| + | <html> | ||
| - | + | <br> | |
| - | + | <table border="0" width="100%" style="background-color:#000000"> | |
| - | + | <tr> | |
| - | + | <th> | |
| - | + | <image src="https://static.igem.org/mediawiki/2010/2/29/UofLteamlogo.jpg" width="200px"/> | |
| - | + | ||
| - | + | </th> | |
| - | + | ||
| - | + | ||
| - | + | <th> | |
| - | + | ||
| - | + | ||
| - | + | <image src="https://static.igem.org/mediawiki/2010/9/91/UofLLabWork.JPG" height="300px"/> | |
| - | + | ||
| - | + | ||
| - | + | </th> | |
| - | + | ||
| - | + | ||
| - | + | <th> | |
| - | + | ||
| - | + | ||
| - | + | <image src="https://static.igem.org/mediawiki/2010/2/29/UofLteamlogo.jpg" width="200px"/> | |
| - | + | ||
| - | + | ||
| - | + | </th> | |
| - | + | ||
| - | + | ||
| - | + | </tr> | |
| - | + | ||
| - | + | ||
| - | + | </table> | |
| - | + | ||
| - | + | ||
| - | + | <br> | |
| - | =July 2010= | + | |
| - | ==July 5/2010== | + | <align="centre"> |
| + | <table border="0" width="100%" style="background-color:#000000"> | ||
| + | |||
| + | <tr> | ||
| + | |||
| + | <th> | ||
| + | |||
| + | <a href="https://2010.igem.org/Team:Lethbridge"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/2/22/UofLHome.jpg" width="80"/> | ||
| + | </a> | ||
| + | |||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Team"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/0d/UofLTeam.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Project"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/8d/UofLProjectbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLNotebookbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Parts"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/84/UofLPartsSubmittedToTheRegistrybutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Modeling"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/e/e1/UofLModelingbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Ethics"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/2/26/UofLEthicsbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Safety"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/00/UofLSafetybutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Art"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/0a/UofLArt.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/News"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/c/c3/UofLNewsButton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | </table> | ||
| + | </body> | ||
| + | </html> | ||
| + | <hr> | ||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <font color="white">Feel free to look around our notebook! | ||
| + | </center> | ||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
| + | <center> | ||
| + | <table border="0" width="28%" style="background-color:#000000"> | ||
| + | |||
| + | <tr> | ||
| + | <th> | ||
| + | <div class="miniBar"> | ||
| + | <div class="countdown"><object type="application/x-shockwave-flash" data="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=10&date_day=27&date_year=0&un=THE WIKI FREEZE&size=normal&mo=10&da=27&yr=2010" width="300" height="100"><param name="movie" value="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=10&date_day=27&date_year=0&un=THE WIKI FREEZE&size=normal&mo=10&da=27&yr=2010" /><param name="bgcolor" value="#000000" /></object><img src="http://www.oneplusyou.com/q/img/bb_badges/countdown.jpg" alt="" style="display: none;" height="1" width="1" /></div> | ||
| + | <div class="miniContainer"> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLNotebookbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Protocols"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/9/91/UofLprotocolsbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Calendar"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLcalendar.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th> | ||
| + | <div class="miniBar"> | ||
| + | <div class="countdown"><object type="application/x-shockwave-flash" data="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=11&date_day=05&date_year=0&un=THE IGEM JAMBOREE&size=normal&mo=11&da=05&yr=2010" width="300" height="100"><param name="movie" value="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=11&date_day=05&date_year=0&un=THE IGEM JAMBOREE&size=normal&mo=11&da=05&yr=2010" /><param name="bgcolor" value="#000000" /></object><img src="http://www.oneplusyou.com/q/img/bb_badges/countdown.jpg" alt="" style="display: none;" height="1" width="1" /></div> | ||
| + | <div class="miniContainer"> | ||
| + | </th> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | <hr> | ||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <font color="white">Here you can check out the work we have done in the lab, click on a month to take a look! | ||
| + | </center> | ||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
| + | <center> | ||
| + | <table border="0" width="50%" style="background-color:#000000"> | ||
| + | |||
| + | <tr> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/April"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/8a/UofLapril.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/May"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/7/7b/UofLmaybutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/June"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/80/UofLjunebutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/July"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/5/53/UofLjulybutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/August"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/1/15/UofLaugustbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/September"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/4/4d/UofLseptemberbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/October"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/4/4e/UofLoctoberbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | |||
| + | |||
| + | <tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | </body> | ||
| + | </html> | ||
| + | <hr> | ||
| + | |||
| + | <BLOCKQUOTE> | ||
| + | |||
| + | =<font color="white">July 2010= | ||
| + | ==<font color="white">July 3/2010== | ||
| + | (In Lab: JS)<br> | ||
| + | |||
| + | <b>Objective:</b> To restrict and ligate pBAD-TetR and YEPs into pSB1C3 backbone.<br> | ||
| + | |||
| + | <b>Method:</b> Used [[Team:Lethbridge/Notebook/Protocols|Restriction of Plasmid DNA]] protocol and ligated the parts into pSB1C3.<br> | ||
| + | |||
| + | ==<font color="white">July 5/2010== | ||
(In Lab: JV, AV, HB)<br> | (In Lab: JV, AV, HB)<br> | ||
| Line 98: | Line 256: | ||
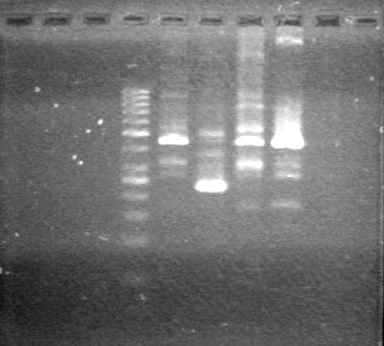
Ran gel at 100V for 45 minutes.<br> | Ran gel at 100V for 45 minutes.<br> | ||
| - | < | + | <b>Results:</b> |
| + | [[image:Lethbridge_100705AVPCR.JPG|200px]]<br> | ||
| - | ==July 5/2010 Evening== | + | ==<font color="white">July 5/2010 Evening== |
<b>Objective:</b> To over-express CFP complete in DH5α<br> | <b>Objective:</b> To over-express CFP complete in DH5α<br> | ||
| Line 109: | Line 268: | ||
2) Went in the shaker at 6:50pm | 2) Went in the shaker at 6:50pm | ||
| - | ==July 6/2010== | + | ==<font color="white">July 6/2010== |
(In lab: JV, AV, HB)<br> | (In lab: JV, AV, HB)<br> | ||
<b>Objective:</b> To continue the over-expression of CFP complete in DH5α<br> | <b>Objective:</b> To continue the over-expression of CFP complete in DH5α<br> | ||
| Line 165: | Line 324: | ||
Ran at 100V for 80 mins.<br> | Ran at 100V for 80 mins.<br> | ||
| - | < | + | <b>Results:</b> |
| + | [[image:Lethbridge_100705AV BW (2).jpg|200px]]<br> | ||
<font color ="Red">More to fill in, but Anthony does not understand the stuff written in the lab book</font><br> | <font color ="Red">More to fill in, but Anthony does not understand the stuff written in the lab book</font><br> | ||
| - | ==July 8/2010== | + | ==<font color="white">July 8/2010== |
(In lab: JV, AV, HB, HS)<br> | (In lab: JV, AV, HB, HS)<br> | ||
<b>Objective:</b><br> | <b>Objective:</b><br> | ||
| - | ==July 8/2010 - Evening== | + | <font color ="red"> Anthony does not understand the stuff written in the lab book.</font> |
| + | |||
| + | ==<font color="white">July 8/2010 - Evening== | ||
(In lab: KG)<br> | (In lab: KG)<br> | ||
<b>Objective:</b>To transform TetR in pSB1A2 plasmid (BBa_C0040 - 2010 iGEM Distribution Kit Plate Well 4A) and pTetR in pSB1A2 plasmid (BBa_R0040 - 2010 iGEM Distribution Kit Plate Well 6) into DH5α.<br> | <b>Objective:</b>To transform TetR in pSB1A2 plasmid (BBa_C0040 - 2010 iGEM Distribution Kit Plate Well 4A) and pTetR in pSB1A2 plasmid (BBa_R0040 - 2010 iGEM Distribution Kit Plate Well 6) into DH5α.<br> | ||
| - | <b>Method:</b>Used the Competent Cell Transformation Protocol as well as transformed pUC19 as a positive control.< | + | <b>Method:</b>Used the Competent Cell Transformation Protocol as well as transformed pUC19 as a positive control.<br> |
<b>Results:</b> | <b>Results:</b> | ||
| Line 190: | Line 352: | ||
</table><br> | </table><br> | ||
| - | ==July 9/2010== | + | ==<font color="white">July 9/2010== |
(In lab: JV)<br> | (In lab: JV)<br> | ||
<b>Objective:</b>To overexpress pLacI-mRBS-mms6-dT construct.<br> | <b>Objective:</b>To overexpress pLacI-mRBS-mms6-dT construct.<br> | ||
| Line 211: | Line 373: | ||
<font color ="red">SDS PAGE picture!!!!!!!!!!!</font> | <font color ="red">SDS PAGE picture!!!!!!!!!!!</font> | ||
| - | ==July 10/2010== | + | ==<font color="white">July 10/2010== |
(In lab: JV)<br> | (In lab: JV)<br> | ||
<b>Objective:</b>To determine if maxipreps finished on July 9th and 10th have significant concentrations of DNA.<br> | <b>Objective:</b>To determine if maxipreps finished on July 9th and 10th have significant concentrations of DNA.<br> | ||
| Line 231: | Line 393: | ||
</table><br> | </table><br> | ||
| - | Ran at 100V for 40 minutes. Strained in EtBr for 10 minutes. | + | Ran at 100V for 40 minutes. Strained in EtBr for 10 minutes.<br> |
| - | ==July 12/2010== | + | <b>Results:</b> |
| + | [[image:Lethbridge_100710JV BW.JPG|200px]] | ||
| + | |||
| + | ==<font color="white">July 12/2010== | ||
(In lab: AV,HB,JV)<br> | (In lab: AV,HB,JV)<br> | ||
| - | <b>Objective:</b> | + | <b>Objective:</b> Maxiprep pLacI (A2) from glycerol stock.<br> |
| + | |||
| + | <b>Method:</b> Used Maxiprep Protocol. Cell pellet weighed 1.02g.<br> | ||
| + | |||
| + | <b>Objective:</b> Add dT to the end of mms6, xylE, and lumazine. | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | 1) Restrict "Part 1" BioBricks: mms6, xylE, and lumazine with EcoRI and SpeI.<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>pDNA</td><td>2</td></tr> | ||
| + | <tr><td>Red Buffer</td><td>2</td></tr> | ||
| + | <tr><td>EcoRI</td><td>0.25</td></tr> | ||
| + | <tr><td>SpeI</td><td>0.25</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>15.5</td></tr> | ||
| + | </table><br> | ||
| + | 2) Restrict "Part 2" BioBrick: dT with EcoRI and XbaI.<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td><td><b>2.5x Volume (µL)</b></td></tr> | ||
| + | <tr><td>pDNA</td><td>2</td><td>5</td></tr> | ||
| + | <tr><td>Orange Buffer</td><td>2</td><td>5</td></tr> | ||
| + | <tr><td>EcoRI</td><td>0.25</td><td>0.625</td></tr> | ||
| + | <tr><td>XbaI</td><td>0.25</td><td>0.625</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>15.5</td><td>38.75</td></tr> | ||
| + | </table><br> | ||
| + | 3) 1% Agarose gel (1x TAE) of restricted DNA | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Component (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>1 kb Ladder</td><td>0.5 ladder + 2 loading dye (6x) + 7.5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>dT</td><td>8 DNA + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>3</td><td>dT restricted</td><td>8 DNA + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>4</td><td>mms6</td><td>8 DNA + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>5</td><td>mms6 restricted</td><td>8 DNA + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>6</td><td>xylE</td><td>8 DNA + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>7</td><td>xylE restricted</td><td>8 DNA + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>8</td><td>Lumazine</td><td>8 DNA + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>9</td><td>Lumazine restricted</td><td>8 DNA + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>10</td><td>Empty</td><td>Empty</td></tr> | ||
| + | </table><br> | ||
| + | Ran at 100V for 43 minutes. Stained in EtBr for 10 minutes.<br> | ||
| + | <b>Results:</b> | ||
| + | [[image:Lethbridge_100712HB REstriction.JPG|200px]]<br> | ||
| + | |||
| + | 4) Ligate restricted dT to the ends of the "Part 1" Biobricks.<br> | ||
| + | |||
| + | ==<font color="white">July 12/2010 - Evening== | ||
| + | (In lab: KG,TF)<br> | ||
| + | <b>Objective:</b> Continue with addition of dT to the ends of mms6, xylE, and lumazine.<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | 4) Ligate restricted dT to the ends of the "Part 1" Biobricks.<br> | ||
| + | dT to mms6:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>T4 DNA Ligase</td><td>0.25</td></tr> | ||
| + | <tr><td>10x T4 Ligation Buffer</td><td>2</td></tr> | ||
| + | <tr><td>dT</td><td>8.3</td></tr> | ||
| + | <tr><td>mms6</td><td>9.2</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>0.25</td></tr> | ||
| + | </table><br> | ||
| + | dT to xylE:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>T4 DNA Ligase</td><td>0.25</td></tr> | ||
| + | <tr><td>10x T4 Ligation Buffer</td><td>2</td></tr> | ||
| + | <tr><td>dT</td><td>8.3</td></tr> | ||
| + | <tr><td>xylE</td><td>3.4</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>6.05</td></tr> | ||
| + | </table><br> | ||
| + | dT to lumazine:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>T4 DNA Ligase</td><td>0.25</td></tr> | ||
| + | <tr><td>10x T4 Ligation Buffer</td><td>2</td></tr> | ||
| + | <tr><td>dT</td><td>8.3</td></tr> | ||
| + | <tr><td>lumazine</td><td>3.35</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>6.1</td></tr> | ||
| + | </table><br> | ||
| + | Ligations were incubated at room temperature overnight.<br> | ||
| + | |||
| + | |||
| + | <b>Objective:</b> Prepare mastermix for four PCR reactions for following day.<br> | ||
| + | |||
| + | <b>Method:</b> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td><td><b>5x Volume (µL)</b></td></tr> | ||
| + | <tr><td>10mM dNTPs</td><td>1</td><td>5</td></tr> | ||
| + | <tr><td>5x Phusion Buffer</td><td>4</td><td>20</td></tr> | ||
| + | <tr><td>Forward Primer (VF2)</td><td>1</td><td>5</td></tr> | ||
| + | <tr><td>Reverse Primer (VR)</td><td>1</td><td>5</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>10.8</td><td>54</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | ==<font color="white">July 13/2010== | ||
| + | (in lab: JV)<br> | ||
| + | <b>Objective:</b> To determine if ligations of previous day (July 12/2010) were successful.<br> | ||
| + | <b>Method:</b><br> | ||
| + | 1) Use prepared mastermix to run a PCR of ligated mms6-dT, ligated xylE-dT, ligated lumazine-dT, and control-dT<br> | ||
| + | - To each PCR tube, add 17.8&mirco;L of mastermix, 0.2µL of Phusion DNA polymerase, and 2µL of template DNA.<br> | ||
| + | - Ran PCR's for 36 cycles using the iGEM preset.<br> | ||
| + | |||
| + | 2) 2% Agarose gel<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Components (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>50 bp Ladder</td><td>1 ladder + 2 loading dye (6x) + 7 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>mms6-dT</td><td>8 PCR product + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>3</td><td>xylE-dT</td><td>8 PCR product + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>4</td><td>lumazine-dT</td><td>8 PCR product + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>5</td><td>dT</td><td>8 PCR product + 2 loading dye (6x)</td></tr> | ||
| + | <tr><td>6</td><td>Empty</td><td>Empty</td></tr> | ||
| + | <tr><td>7</td><td>Empty</td><td>Empty</td></tr> | ||
| + | <tr><td>8</td><td>Empty</td><td>Empty</td></tr> | ||
| + | <tr><td>9</td><td>Empty</td><td>Empty</td></tr> | ||
| + | <tr><td>10</td><td>Empty</td><td>Empty</td></tr> | ||
| + | </table><br> | ||
| + | Ran at 100V for __ minutes. Stained in EtBr for 10 minutes.<br> | ||
| + | |||
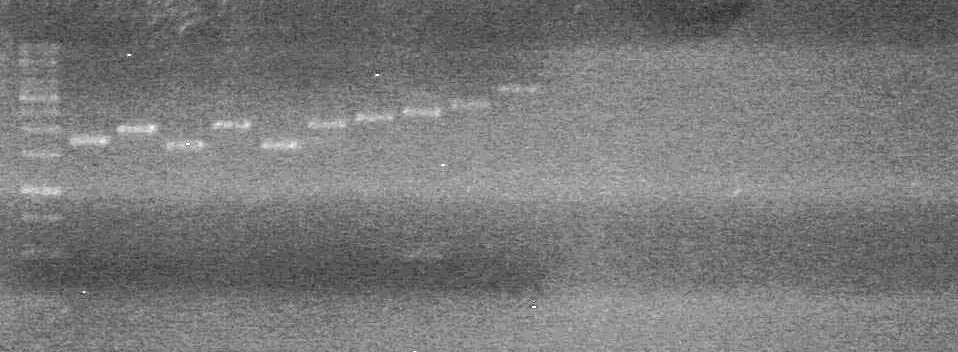
| + | <b>Results:</b> | ||
| + | [[image:Lethbridge_100713JV PCR.JPG|200px]]<br> | ||
| + | |||
| + | ==<font color="white">July 14/2010== | ||
| + | (in lab:J.S, K.G )<br> | ||
| + | <b>Objective:</b> Inoculate culture for maxi prep with placI | ||
| + | |||
| + | <b>Method:</b> | ||
| + | To a 450mL solution of LB media 4.5(µL) of ampicillin was added with glycerol placI aseptically. | ||
| + | ---- | ||
| + | (in lab:J.S, K.G )<br> | ||
| + | <b>Objective:</b> Restriction of dT and mms6 | ||
| + | |||
| + | <b>protocol</b> | ||
| + | |||
| + | 1) | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>Buffer&*</td><td>2</td></tr> | ||
| + | <tr><td>10x T4 Milli Q H<sub>2</sub>O</td><td>15.5</td></tr> | ||
| + | <tr><td>pDNA**</td><td>2</td></tr> | ||
| + | <tr><td>Restriction enzymes***</td><td>0.25</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | <b>*</b> Orange buffer was used for dT, and Red buffer was used for mms6 | ||
| + | |||
| + | <b>**</b>pDNA was dT and mms6 | ||
| + | |||
| + | <b>***</b>Restriction enzymes for dT were XbalI and EcoRI, and for mms6 SpeI and EcoRI | ||
| + | |||
| + | 2) | ||
| + | Reaction was incubated at 37<sup>0</sup>C for 1 hour. Start 8:50pm till 9:50pm | ||
| + | After incubation reaction was heat shocked at 80<sup>0</sup>C for 20 minutes | ||
| + | |||
| + | ---- | ||
| + | (in lab:J.S, K.G )<br> | ||
| + | <b>Objective:</b> Ligation of dT to each of mms6, xylE and lumazine. | ||
| + | |||
| + | <b>protocol</b> | ||
| + | |||
| + | 1) | ||
| + | |||
| + | dT to mms6:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>T4 DNA Ligase</td><td>0.25</td></tr> | ||
| + | <tr><td>10x T4 Ligation Buffer</td><td>2</td></tr> | ||
| + | <tr><td>dT</td><td>8.3</td></tr> | ||
| + | <tr><td>mms6</td><td>9.2</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>0.25</td></tr> | ||
| + | </table><br> | ||
| + | dT to xylE:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>T4 DNA Ligase</td><td>0.25</td></tr> | ||
| + | <tr><td>10x T4 Ligation Buffer</td><td>2</td></tr> | ||
| + | <tr><td>dT</td><td>8.3</td></tr> | ||
| + | <tr><td>xylE</td><td>3.4</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>6.05</td></tr> | ||
| + | </table><br> | ||
| + | dT to lumazine:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>T4 DNA Ligase</td><td>0.25</td></tr> | ||
| + | <tr><td>10x T4 Ligation Buffer</td><td>2</td></tr> | ||
| + | <tr><td>dT</td><td>8.3</td></tr> | ||
| + | <tr><td>lumazine</td><td>3.35</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>6.1</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | 2) | ||
| + | Incubated reaction overnight at room temperature<br> | ||
| + | |||
| + | ==<font color="white">July 15/2010== | ||
| + | (in lab: AV)<br> | ||
| + | <b>Objective:</b> Maxiprep pLacI and mms6.<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>component</b></td><td><b>pallet weight (g)</b></td></tr> | ||
| + | <tr><td>mms6</td><td>1.02</td></tr> | ||
| + | <tr><td>pLacI</td><td>1.54</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | ==<font color="white">July 15/2010 Evening== | ||
| + | (in lab: AV)<br> | ||
| + | <b>Objective:</b> transform ligations from July 14 and July 12,2010;xylE/dt, mms6/dt lumazine/dt into DH5&alpha. | ||
| + | Also to transform mms6 from July 6 and July 10 into Bl21(DE3) | ||
| + | |||
| + | <b>Protocol:</b> | ||
| + | to transform competent cells see protocol:[https://2010.igem.org/Team:Lethbridge/Notebook/Protocols#Competent_Cell_Transformation] | ||
| + | |||
| + | <b>*</b> cells were incubated on ice for 30 minutes started at 7:00-7:30. | ||
| + | <b>**</b>incubated for 1 hour from 8:00 till 9:00 | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>'''Results'''</b></tr> | ||
| + | <tr><td><b>plate</b></td><td><b># of colonies</b></td><td><b>Components (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>200µL xylE-dt July 12</td><td>0</td></tr> | ||
| + | <tr><td>2</td><td>200µL mms6-dt July 12</td><td>1</td></tr> | ||
| + | <tr><td>3</td><td>200µL lumazine-dt</td><td>1</td></tr> | ||
| + | <tr><td>4</td><td>200µL mms6 maxiprep July 6</td><td>Lawn</td></tr> | ||
| + | <tr><td>5</td><td>200µL positve control puC19 into BL21(DE3)</td><td>Lawn</td></tr> | ||
| + | <tr><td>6</td><td>200µL xylE-dt July 14</td><td>0</td></tr> | ||
| + | <tr><td>7</td><td>200µL mms6-dt July 14</td><td>120</td></tr> | ||
| + | <tr><td>8</td><td>200µL lumazine-dt July14</td><td>0</td></tr> | ||
| + | <tr><td>9</td><td>200µL mms6 maxiprep July 10</td><td>.</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | <b>*</b>because of a shortage of plates not all transformations were plated at 50µL and 200µL | ||
| + | |||
| + | ==<font color="white">July 16,2010== | ||
| + | (in lab: M.C, D.M)<br> | ||
| + | <b>Objective:</b> PCR amplify mms6-dT, xylE-dT, lumazine-dT legations along with dT maxi prep for comparison | ||
| + | |||
| + | <b>Protocol:</b> | ||
| + | |||
| + | Master Mix | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<</td><td>54</td></tr> | ||
| + | <tr><td>Pfu Buffer + MgSO<sub>4</sub></td><td>20</td></tr> | ||
| + | <tr><td>10 mM dMTPs</td><td>5</td></tr> | ||
| + | <tr><td>forward primer</td><td>5</td></tr> | ||
| + | <tr><td>reverse primer</td><td>5</td></tr> | ||
| + | </table><br> | ||
| + | 89 µL TOTAL--> 17.8 into each PCR reaction | ||
| + | |||
| + | + 2 µL ligation | ||
| + | + 2 µL Pfu polymerase | ||
| + | |||
| + | Ran iGem-ligTest in thermocycler | ||
| + | |||
| + | |||
| + | ==<font color="white">July 19, 2010== | ||
| + | (in lab: A.V, H.B)<br> | ||
| + | <b>Objective:</b> Purification pf pLacI and mms6 maxipreps done on July 14 and 16, 2010 using the biobasic protocol for purification of PCR products. | ||
| + | |||
| + | <b>Objective:</b> Add pLacI to sRBS and add dT to xylE and lumazine | ||
| + | |||
| + | <b>Method:</b> | ||
| + | |||
| + | Restrict pLacI, xylE and lumazine with SpeI and EcoRI and restrict dT and sRBS with EcorI and XbaI | ||
| + | |||
| + | 1)Restrictions: | ||
| + | |||
| + | pLacI, xylE, and Lumazine | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O</td><td>15.5</td></tr> | ||
| + | <tr><td>Red Buffer<sub>4</sub></td><td>2</td></tr> | ||
| + | <tr><td>SpeI</td><td>0.25</td></tr> | ||
| + | <tr><td>EcoRI</td><td>0.25</td></tr> | ||
| + | <tr><td>pDNA</td><td>2</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | dT | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O</td><td>38.75</td></tr> | ||
| + | <tr><td>Orange Buffer<sub>4</sub></td><td>5</td></tr> | ||
| + | <tr><td>XbaI</td><td>0.625</td></tr> | ||
| + | <tr><td>EcoRI</td><td>0.625</td></tr> | ||
| + | <tr><td>pDNA</td><td>5</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | sRBS | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O</td><td>15.5</td></tr> | ||
| + | <tr><td>Red Buffer<sub>4</sub></td><td>2</td></tr> | ||
| + | <tr><td>xBal</td><td>0.25</td></tr> | ||
| + | <tr><td>EcoRI</td><td>0.25</td></tr> | ||
| + | <tr><td>pDNA</td><td>2</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | Restriction incubation at 37.5<sup>0</sup>C started at 11:55am. Ended at 12:55pm | ||
| + | Heat killed enzymes at 80<sup>0</sup>C for 20 minutes | ||
| + | |||
| + | 2)Restrictions were run on a 1% agarose gel (1 X TAE) | ||
| + | 1% Agarose gel<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Components (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>1kB Ladder</td><td>0.5 ladder + 2 loading dye (5x) + 5.5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>pLacI</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | <tr><td>3</td><td>pLacI restriction digest</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | <tr><td>4</td><td>sRBS</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | <tr><td>5</td><td>sRBS restriction digest</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | <tr><td>6</td><td>xylE</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | <tr><td>7</td><td>xylE restriction digest</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | <tr><td>8</td><td>lumazine</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | <tr><td>9</td><td>lumazine restriction digest</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | <tr><td>10</td><td>dT</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | <tr><td>11</td><td>dT restriction digest</td><td>6 pDNA + 2 loading dye (5x)</td></tr> | ||
| + | </table><br> | ||
| + | |||
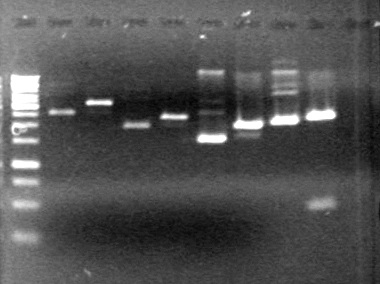
| + | Gel ran for 70 minutes at 100V and was stained in EtBr for 10 minutes | ||
| + | |||
| + | <b>Results:</b> | ||
| + | [[image:Lethbridge_100719HBAVDigest.JPG|200px]]<br> | ||
| + | |||
| + | Results after quantifying restriction digest | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Content</b></td><td><b> Quantity of DNA (ng/µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>1kB Ladder</td><td>12.5</td></tr> | ||
| + | <tr><td>3</td><td>pLacI restriction digest</td><td>5.21</td></tr> | ||
| + | <tr><td>5</td><td>sRBS restriction digest</td><td>3.72</td></tr> | ||
| + | <tr><td>7</td><td>xylErestriction digest</td><td>3.36</td></tr> | ||
| + | <tr><td>9</td><td>Lumazine restriction digest</td><td>2.63</td></tr> | ||
| + | <tr><td>11</td><td>dT restriction digest</td><td>2.98</td></tr> | ||
| + | </table><br> | ||
| + | |||
| + | ==<font color="white">July 19, 2010 Evening== | ||
| + | (K.G)<br> | ||
| + | <b>Objective:</b> Ligate together rbs-xylE and dT, lumazine and dT, also pLacI and sRBS.<br> | ||
| + | |||
| + | <b>Method:</b> All ligation mixes had:<br> | ||
| + | *10X T4 Ligation Buffer | ||
| + | *Milli-Q H<sub>2</sub>O to fill to 20µL | ||
| + | *T4 DNA Ligase | ||
| + | *20ng plasmid DNA<br> | ||
| + | |||
| + | Ligations were left over-night at room temperature. <br> | ||
| + | |||
| + | (J.V.)<br> | ||
| + | <b>Objective:</b>Determine the results of the transformations done by K.G. on July 15/2010.<br> | ||
| + | |||
| + | <b>Method:</b> Inoculate 5mL LB media w/Amp and colony. Incubated over night at 37<sup>o</sup>C.<br> | ||
| + | |||
| + | <b>Results:</b> Lumazine Synthase with dT ligated onto it grew.<br> | ||
| + | |||
| + | ==<font color="white">July 20, 2010== | ||
| + | (AV, HB)<br> | ||
| + | <b>Objective:</b>Miniprep lumazine-dt and 4 N-terminus tags and analyze.<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | *Use [[Team:Lethbridge/Notebook/Protocols|boiling lysis miniprep]] to isolate plasmid DNA. | ||
| + | *PCR amplify BioBrick part to determine if the correct DNA was isolated. | ||
| + | *Ran a 1% Agarose gel to visualize PCR products. | ||
| + | |||
| + | <b><font color="white">Results:</font></b> | ||
| + | Agarose gel did not show any bands.<br> | ||
| + | |||
| + | ==<font color="white">July 20, 2010 Evening== | ||
| + | (AV, HB)<br> | ||
| + | <b>Objective:</b>Transform ligation done on July 19, 2010 in to competent DH5α cells.<br> | ||
| + | * xylE-dT | ||
| + | *lumazine synthase-dT | ||
| + | *pLacI-sRBS | ||
| + | *EYFP and ECFP<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | *Use [[Team:Lethbridge/Notebook/Protocols|Competent Cell Transformation]] | ||
| + | |||
| + | <b>Results:</b><br> | ||
| + | *xylE-dT no colonies | ||
| + | *pLacI-sRBS | ||
| + | *EYFP | ||
| + | *lumazine-dT | ||
| + | *ECFP | ||
| + | |||
| + | ==<font color="white">July 21, 2010== | ||
| + | (JV)<Br> | ||
| + | <b>Objective:</b> Isolate plasmid DNA from pTet, TetR, pET-28(a). | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | *Used [[Team:Lethbridge/Notebook/Protocols|Maxiprep]] protocol. | ||
| + | |||
| + | (JV)<br> | ||
| + | |||
| + | <b>Objective:</b> To induce over-expression of pLacI-RBS-Mms6-dt in BL21(DE3) cells.<br> | ||
| + | |||
| + | <b>Method:</b> Used [[Team:Lethbridge/Notebook/Protocols|Overexpression]].<br> | ||
| + | Ran SDS gel for 78 minutes at 200V<br> | ||
| + | |||
| + | ==<font color="white">July 21, 2010 Evening== | ||
| + | (TF, AS)<br> | ||
| + | <b>Objective:</b> Colony PCR to test for insertions of BioBrick construction, and for quality control of parts received from the Registry. Also to prepare DNA to be sent away for sequencing.<br> | ||
| + | <b>Method:</b><br> | ||
| + | -Ran PCR's<br> | ||
| + | -Ran 2% Agarose gel for:.<br> | ||
| + | *K249001<br> | ||
| + | *K249004<br> | ||
| + | *K249005<br> | ||
| + | *K249006<br> | ||
| + | *K249008<br> | ||
| + | *K249014<br> | ||
| + | *K249017<br> | ||
| + | *mms6-dt<br> | ||
| + | *lumazine-dt<br> | ||
| + | *ECFP<br> | ||
| + | *EYFP<br> | ||
| + | *N-term tag<br> | ||
| + | *xylE-dt<br> | ||
| + | *SRBS-pLacI<br> | ||
| + | *dt<br> | ||
| + | *pTet maxiprep<br> | ||
| + | *pET-28(a) maxiprep<br> | ||
| + | *TetR maxiprep<br> | ||
| + | Gel ran at 100V for 60minutes.<br> | ||
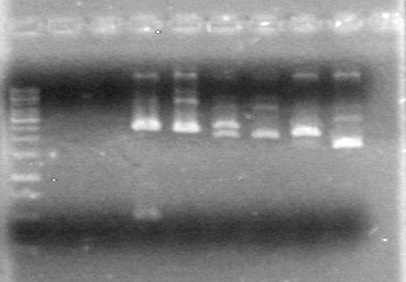
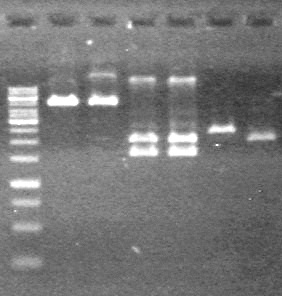
| + | <b>Results:</b> The PCR's did not work. However, the maxipreps show DNA<br> | ||
| + | [[image:Lethbridge_100722 JV BW.jpg|200px]] | ||
| + | |||
| + | ==<font color="white">July 23, 2010== | ||
| + | (JV)<br> | ||
| + | <b>Objective:</b> Insert xylE, Mms6, and lumazine synthase into pET-28(a).<br> | ||
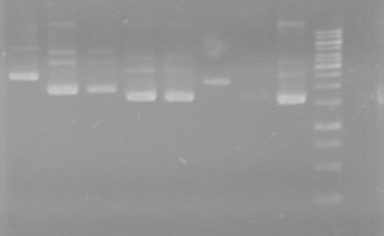
| + | <b>Method:</b> A restriction digest was performed on xylE and Mms6 and ran for 90 minutes at 37 C<br> | ||
| + | <b>Results:</b>Did not see any cut out biobricks.<br> | ||
| + | [[image:Lethbridge_100723MaxiprepRestrictions.jpg|200px]] | ||
| + | |||
| + | ==<font color="white">July 27, 2010== | ||
| + | (in lab: JV, AV, HB)<br> | ||
| + | |||
| + | <b>Objective:</b> To miniprep the overnight cultures of the parts ordered from the Parts Registry and to test the efficiency of the Qiagen Miniprep Kit.<br> | ||
| + | |||
| + | <b>Method:</b> <br> | ||
| + | The following parts were miniprep'd using [[Team:Lethbridge/Notebook/Protocols|Boiling Lysis Plasmid Preparation]]:<br> | ||
| + | *dT (control) | ||
| + | *E0020 (ECFP) | ||
| + | *E0030 (EYFP) | ||
| + | *K249001 | ||
| + | *K249004 | ||
| + | *K249005 | ||
| + | *K249006 | ||
| + | *K249008 | ||
| + | *K249014 | ||
| + | *K249017 | ||
| + | |||
| + | The following parts were miniprep'd using Qiagen Miniprep Kit:<br> | ||
| + | *K249005 | ||
| + | *K249008 | ||
| + | |||
| + | <b>Results:</b> Qiagen Miniprep Kit produced a comparable quantity of DNA to [[Team:Lethbridge/Notebook/Protocols|Boiling Lysis Plasmid Preparation]]. Since the Qiagen Miniprep Kit is faster and easier, all minipreps will be done using the Qiagen Miniprep Kit. | ||
| + | |||
| + | ==<font color="white">July 28, 2010== | ||
| + | (in lab: JV)<br> | ||
| + | |||
| + | <b>Objective:</b> To create a large quantity of pSB1C3 plasmid.<br> | ||
| + | |||
| + | <b>Method:</b> PCR amplified pSB1C3 received from iGEM headquarters. | ||
| + | |||
| + | |||
| + | |||
| + | ==<font color="white">July 29, 2010== | ||
| + | (in lab: HB, AV)<br> | ||
| + | |||
| + | <b>Objective:</b> To PCR amplify the minipreps on July 27, 2010.<br> | ||
| + | |||
| + | <b>Method:</b> PCR amplified the 11 minipreps and dT control.<br> | ||
| + | |||
| + | <b>Results:</b> PCR amplification was successful and 0.2 µL of polymerase will be used per PCR reaction in the future.<br> | ||
| + | |||
| + | <br><br> | ||
| + | <b>Objective:</b> To maxiprep EYFP (E0030), ECFP (E0020), and Lumazine Synthase (K249002).<br> | ||
| + | |||
| + | <b>Method:</b> Used [[Team:Lethbridge/Notebook/Protocols|Maxiprep]]<br> | ||
| + | |||
| + | <table><table border="3"> | ||
| + | <tr><td><b>Cell Pellet</b></td><td><b>Weight (g)</b></td></tr> | ||
| + | <tr><td>ECFP</td><td>2.32</td></tr> | ||
| + | <tr><td>EYFP</td><td>2.00</td></tr> | ||
| + | <tr><td>Lumazine Synthase</td><td>2.66</td></tr></table> | ||
| + | |||
| + | ==<font color="white">July 29, 2010 Evening== | ||
| + | (in lab: KG)<br> | ||
| + | |||
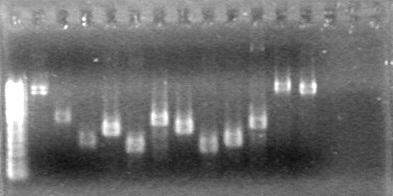
| + | <b>Objective:</b> Run PCR products from the morning on a 2% agarose gel.<br> | ||
| + | |||
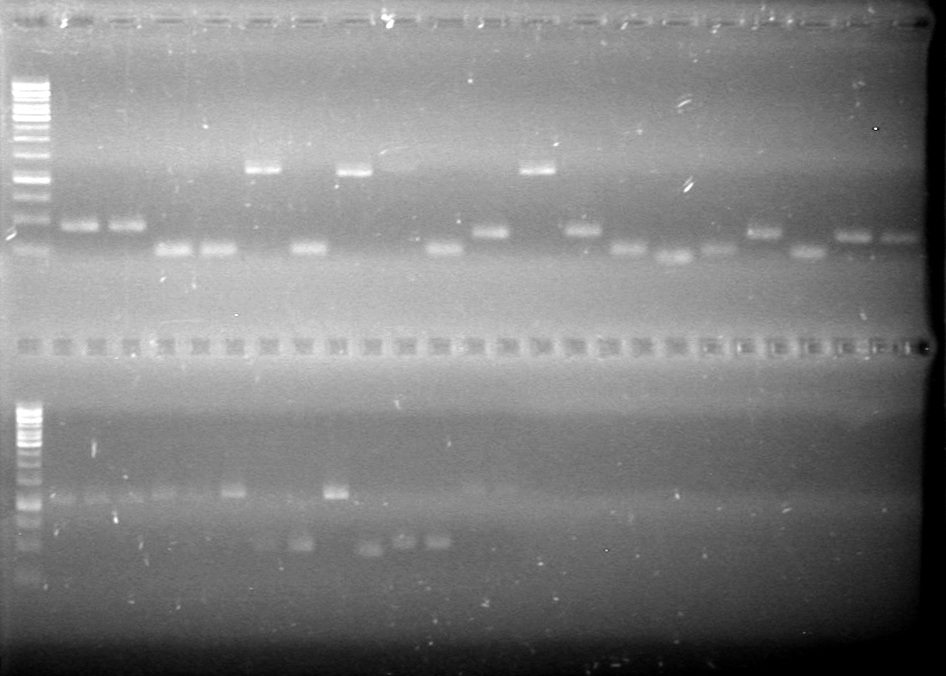
| + | <b>Method:</b> Ran 2% agarose gel of the 11 minipreps and dT control and stained in EtBr for 17 minutes.<br> | ||
| + | <b>Results:</b>PCR amplification of last year's parts not consistent with sizes listed on registry.<br> | ||
| + | [[image:Lethbridge_100729 Registry Parts PCR KG.jpg|200px]]<br><br> | ||
| + | <br> | ||
 "
"