Team:Baltimore US/Project
From 2010.igem.org
m (→Developing low-cost alternatives to existing enzymes: Taq polymerase Project Details: copyediting) |
m (→Developing low-cost alternatives to existing enzymes: Taq polymerase Project Details) |
||
| (One intermediate revision not shown) | |||
| Line 24: | Line 24: | ||
==Developing low-cost alternatives to existing enzymes: ''Taq'' polymerase Project Details== | ==Developing low-cost alternatives to existing enzymes: ''Taq'' polymerase Project Details== | ||
| + | We wished to insert Taq Polymerase into a standard BioBrick vector. If this part should prove useful to other teams as an element of a rational design, we must ensure that no sites for the standard BioBrick restriction enzymes exist within the part itself, otherwise the part would shear upon assembly. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | ====Problem: PstI restriction site | + | We examined the Taq [http://www.ncbi.nlm.nih.gov/nuccore/155128 sequence]and exported the SEQ into Plasma DNA. [http://research.med.helsinki.fi/plasmadna/ Plasma DNA] is free software from University of Helsinki which provides quick analysis of plasmid sequence information. In particular, we obtained a restriction map which identified potential EcoRI, Xbe1, Sbe1, or Pst1 sites within the coding sequence. Here we encountered our first difficulty. |
| - | CTGCAG | + | |
| - | GACGTC | + | ====Problem: a PstI restriction site within the coding sequence==== |
| - | + | At 1717nt, we discovered a restriction site for Pst1: | |
| + | ...CTGCAG... PstI restriction site<br> | ||
| + | ...GACGTC... Complement<br> | ||
| + | |||
| + | We attempted to eliminate the restriction site by employing a site-specific mutagenesis by overlap extension protocol (see [http://www.cshlpress.com/default.tpl?cart=1279686078181232350&fromlink=T&linkaction=full&linksortby=oop_title&--eqSKUdatarq=21 Sambrook, Joseph; Russell, David W. ; Molecular Cloning: A Laboratory Manual, 3rd Edition]). | ||
<br><br> | <br><br> | ||
| - | We then used the Gene Designer 2.0 from [https://www.dna20.com/genedesigner2/ DNA2.0] to analyze the open reading frames and examine the codons within the PstI restriction site. We find that the first three | + | We then used the Gene Designer 2.0 from [https://www.dna20.com/genedesigner2/ DNA2.0] to analyze the open reading frames and examine the codons within the PstI restriction site. We find that the first three code for leucine with CTG; we can substitute the final base pair to yield CTT without sacrificing functional integrity in the manufactured enzyme.<br> |
====Primer Design==== | ====Primer Design==== | ||
| - | We designed a primer pair in order to induce point-mutagenesis at the Pst1 restriction site, flanking the base pair to be altered by 14 nt | + | We designed a primer pair in order to induce point-mutagenesis at the Pst1 restriction site, flanking the base pair to be altered by 14 nt: |
| + | |||
GTGGAGAAGATCCT(T)CAGTACCGGCGG<br> | GTGGAGAAGATCCT(T)CAGTACCGGCGG<br> | ||
CACCTCTTCTAGGA(A)GTCATGGCCGCC<br> | CACCTCTTCTAGGA(A)GTCATGGCCGCC<br> | ||
| - | While we | + | |
| - | + | While we designed the point-mutagenesis primers, we took the opportunity to design and order the primers for the BioBrick Suffix and Prefix. We followed the examples laid out in the Registry of Standard Parts for designing the oligos needed to make a part. | |
| - | Important considerations are | + | Important considerations are melting point and CG concentration, as well as self-dimerizations and hairpins. We analyzed these primers using the [http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/ OligoAnalyzer] from [http://www.idtdna.com/Home/Home.aspx IDT]. When analyzing PolI, only the coding seuence itself was used for sequence inquiry, not the BioBrick Suffix/Prefixes.<br> |
<br> | <br> | ||
====PolI Coli Primers For Overlap Extension PCR==== | ====PolI Coli Primers For Overlap Extension PCR==== | ||
Latest revision as of 03:14, 28 October 2010
| Home | Team | Official Team Profile | Project | Submitted Parts | Modeling | Notebook | Meeting/Lab Times | Safety |
|---|
|
DIY-GEM: a path towards low cost high throughput gene synthesis.Synthetic biology research requires more cost effective approaches toward reagents and hardware accessibility. We are developing low-cost alternatives to existing hardware and enzymes in an attempt to expand participation in biological research and development. Our project expands the accessibility of Taq Polymerase by engineering it in a form compatible with BioBrick assembly. This allows use of the over-expressed enzyme from a crude bacterial extract in a PCR reaction at a fraction of the cost of highly purified commercial enzyme. In addition, we have developed inexpensive and easily assembled lab equipment such as a gel electrophoresis apparatus and a PCR thermal cycler. Enabling researchers to synthesize their own enzymes and having access to inexpensive tools will allow for increased participation among the DIY-bio community, stretch increasingly scarce educational funds, and allow rapid scale up of large scale gene synthesis projects." Developing low-cost alternatives to existing enzymes: Taq polymerase Project DetailsWe wished to insert Taq Polymerase into a standard BioBrick vector. If this part should prove useful to other teams as an element of a rational design, we must ensure that no sites for the standard BioBrick restriction enzymes exist within the part itself, otherwise the part would shear upon assembly.
Problem: a PstI restriction site within the coding sequenceAt 1717nt, we discovered a restriction site for Pst1: ...CTGCAG... PstI restriction site We attempted to eliminate the restriction site by employing a site-specific mutagenesis by overlap extension protocol (see [http://www.cshlpress.com/default.tpl?cart=1279686078181232350&fromlink=T&linkaction=full&linksortby=oop_title&--eqSKUdatarq=21 Sambrook, Joseph; Russell, David W. ; Molecular Cloning: A Laboratory Manual, 3rd Edition]).
Primer DesignWe designed a primer pair in order to induce point-mutagenesis at the Pst1 restriction site, flanking the base pair to be altered by 14 nt: GTGGAGAAGATCCT(T)CAGTACCGGCGG While we designed the point-mutagenesis primers, we took the opportunity to design and order the primers for the BioBrick Suffix and Prefix. We followed the examples laid out in the Registry of Standard Parts for designing the oligos needed to make a part.
Important considerations are melting point and CG concentration, as well as self-dimerizations and hairpins. We analyzed these primers using the [http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/ OligoAnalyzer] from [http://www.idtdna.com/Home/Home.aspx IDT]. When analyzing PolI, only the coding seuence itself was used for sequence inquiry, not the BioBrick Suffix/Prefixes. PolI Coli Primers For Overlap Extension PCRPCR Reaction 1
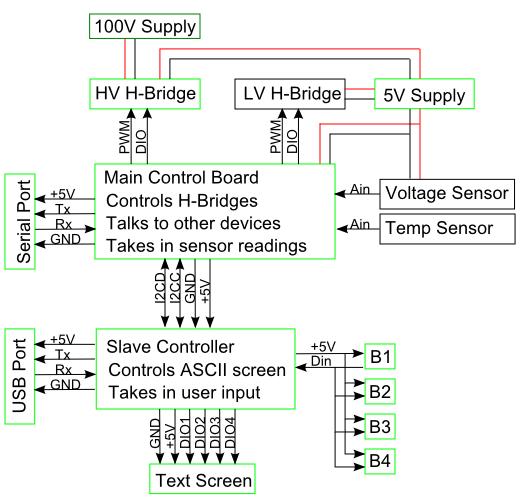
Developing low-cost alternatives to existing hardware: Project Details and ResultsAn unfortunate fact of reality is that precision lab equipment is very costly. Even simple devices such as an Electrophoresis or PCR have significant cost. To ameliorate this a portion of our project will involve designing biological tools that are easy to build and are economical.
|
 "
"