Team:Tokyo Metropolitan/Project/Pattern/Protocol
From 2010.igem.org
(Difference between revisions)
(→About Sample) |
(→About Sample) |
||
| (4 intermediate revisions not shown) | |||
| Line 292: | Line 292: | ||
<table border="1" align="center"> | <table border="1" align="center"> | ||
| - | <TR><TD> No. </TD><TD></TD> | + | <TR><TD> No. </TD><TD>template</TD><TD>'''fragment'''<TD>Digest enzyme</TD><TD>bp<TR> |
| - | <TR><TD> 1 </TD><TD>BBa_K208017</TD><TD>Promoter-signal</TD><TD>AvrⅡ</TD><TD>266<TR> | + | <TR><TD> 1 </TD><TD>BBa_K208017</TD><TD>'''Promoter-signal'''</TD><TD>AvrⅡ</TD><TD>266<TR> |
| - | <TR><TD> 2 </TD><TD> | + | <TR><TD> 2 </TD><TD>E.coli genome</TD><TD>'''CyaA'''</TD><TD>NheⅠ</TD><TD>2583<TR> |
| - | <TR><TD> 3 </TD><TD>BBa_l12521</TD><TD>mRFP1-terminator</TD><TD>NheⅠ</TD><TD>861<TR> | + | <TR><TD> 3 </TD><TD>BBa_l12521</TD><TD>'''mRFP1-terminator'''</TD><TD>NheⅠ</TD><TD>861<TR> |
| - | <TR><TD> 4 </TD><TD>BBa_K208017</TD><TD>Promoter</TD><TD>AvrⅡ</TD><TD>145<TR> | + | <TR><TD> 4 </TD><TD>BBa_K208017</TD><TD>'''Promoter'''</TD><TD>AvrⅡ</TD><TD>145<TR> |
| - | <TR><TD> 5 </TD><TD>BBa_K208017</TD><TD>RBS-signal</TD><TD>SpeⅠ</TD><TD>103<TR> | + | <TR><TD> 5 </TD><TD>BBa_K208017</TD><TD>'''RBS-signal'''</TD><TD>SpeⅠ</TD><TD>103<TR> |
| - | <TR><TD> 6 </TD><TD>BBa_l732901</TD><TD>LacZ</TD><TD>SpeⅠ</TD><TD>3093<TR> | + | <TR><TD> 6 </TD><TD>BBa_l732901</TD><TD>'''LacZ'''</TD><TD>SpeⅠ</TD><TD>3093<TR> |
| - | <TR><TD> 7 </TD><TD>BBa_l12521</TD><TD>terminator</TD><TD>NheⅠ</TD><TD>167<TR> | + | <TR><TD> 7 </TD><TD>BBa_l12521</TD><TD>'''terminator'''</TD><TD>NheⅠ</TD><TD>167<TR> |
| - | <TR><TD> 8 </TD><TD> | + | <TR><TD> 8 </TD><TD>E. coli genome</TD><TD>'''CRP'''</TD><TD>AvrⅡ</TD><TD>657<TR> |
| - | <TR><TD> 9 </TD><TD>BBa_K156010</TD><TD>SBFP2</TD><TD>XbaⅠ</TD><TD>721<TR> | + | <TR><TD> 9 </TD><TD>BBa_K156010</TD><TD>'''SBFP2'''</TD><TD>XbaⅠ</TD><TD>721<TR> |
| - | <TR><TD> 10 </TD><TD>BBa_J23110</TD><TD>Promoter-RBS</TD><TD>SpeⅠ</TD><TD>70<TR> | + | <TR><TD> 10 </TD><TD>BBa_J23110</TD><TD>'''Promoter-RBS'''</TD><TD>SpeⅠ</TD><TD>70<TR> |
</table> | </table> | ||
Latest revision as of 10:03, 27 October 2010

PCR with Pho DNA Polymerase (NIPPON GENE)
<Materials>
- 10x reaction buffer
- Forward primer (starting with 20µM)
- Reverse primer (starting with 20µM)
- Pho DNA Polymerase (starting with 2.5U/µl)
- dNTP (starting with 2.5mM)
- DW
- DNA samples
<Process>
- Make the pellet with pre-colony
- Thaw all required reagents completely and put them on ice. Mix all reagents well by inversion and spin them down prior to pipeting.
- Prepare the reaction mix to correct for dispensing losses prepare an excess of reaction mix (for example, a 100 reactions mix for 96 reactions)
- 10x reaction buffer 12.5μl
- Forward primer 2.5μl (starting with 20µM)
- Reverse primer 2.5μl (starting with 20µM)
- Pho DNA Polymerase 2.5µl (starting with 2.5U/µl)
- dNTP 20μl (starting with 2.5mM)
- DW 85µl
- Total reaction Mix 125μl
- 10x reaction buffer 12.5μl
- Add all components together, except for the template. Mix thoroughly by inversion. Spin down.
- Add the template DNA(pellet or Plasmid) for your samples in to PCR tubes
- Add 50μl of the reaction mix per tube and mix gently on a stirrer or spin down. Ensure that no bubbles are present in the reaction tube. Reaction set up can be done at room temperature.
- Program your Real-Time Thermocycler w/ “fast block” using the following recommended FAST parameters:
- 95°C 5min.
- 95°C 30sec. 30cycle
- 55°C 30sec. 30cycle
- Tm-5°C 2~4min. 30cycle
- 72°C 5min.
- 4°C ∞
- 95°C 5min.
PCR with Tag DNA Polymerase (NIPPON GENE)
<Materials>
- 10x reaction buffer
- Forward primer (starting with 20µM)
- Reverse primer (starting with 20µM)
- Tag DNA Polymerase (starting with 2.5U/µl)
- dNTP (starting with 2.5mM)
- DW
- DNA samples
<Process>
- Make the pellet with pre-culture
- Thaw all required reagents completely and put them on ice. Mix all reagents well by inversion and spin them down prior to pipeting.
- Prepare the reaction mix to correct for dispensing losses prepare an excess of reaction mix (for example, a 100 reactions mix for 96 reactions)
- 10x reaction buffer 12.5μl
- Forward primer 2.5μl (starting with 20µM)
- Reverse primer 2.5μl (starting with 20µM)
- Tag DNA Polymerase 1.0µl (starting with 2.5U/µl)
- dNTP 10μl (starting with 2.5mM)
- DW 96.5µl
- Total reaction Mix 125μl
- 10x reaction buffer 12.5μl
- Add all components together, except for the template. Mix thoroughly by inversion. Spin down.
- Add the template DNA(pellet or Plasmid) for your samples in to PCR tubes
- Add 50μl of the reaction mix per tube and mix gently on a stirrer or spin down. Ensure that no bubbles are present in the reaction tube. Reaction set up can be done at room temperature.
- Program your Real-Time Thermocycler w/ “fast block” using the following recommended FAST parameters:
- 95°C 5min.
- 95°C 30sec. 30cycle
- 55°C 30sec. 30cycle
- Tm-5°C 2~4min. 30cycle
- 72°C 5min.
- 4°C ∞
- 95°C 5min.
DNA Ligation with Ligation-convenience kit (NIPPON GENE)
<Materials>
- DNA solution
- 2 × Ligation Mix (Ligation-convenience kit from NIPPON GENE)
<Process>
- Prepare 10 μl of DNA solution to contain DNA fragment with appropriate mole ratio against vector DNA.
- Add 10μl of 2 × Ligation Mix to the DNA solution and mix well.
- Ligation reaction. Incubate 5-30min at 16°C.
- Apply DNA reaction mixture directly to transformation or in vitro packaging as it is.
DNA extraction
<Materials>
- Binding Buffer (5M Guanidine Thiocyanate; 100mM Tris-HCl (pH7.0))
- Wash Buffer (10mM Tris-HCl (pH7.5))
- Silica gel solution
- TE (10mM Tris-HCl pH 8.0; 0.1mM EDTA)
- Sample
<Process>
- Add 150µl of Binding Buffer to the tube and Vortex.
- Add 10µl of Silica gel solution to the tube and Vortex.
- Vortex every 1min for 5min.
- Centrifuge on 12,000 rpm for 1min, and then aspirate the supernatant.
- Add 200µl of Wash Buffer to tube and Vortex.
- Centrifuge on 12,000 rpm for 30sec, and then aspirate the supernatant.
- Add 200µl of Wash Buffer to tube and Vortex.
- Centrifuge on 12,000 rpm for 30sec, and then aspirate the supernatant.
- Dry the pellets in the speed-vac for 5min, or air dry.
- Add 20µl of TE Buffer to the dried pellets
DNA extraction with ISOPLANT(NIPPON GENE)
<Materials>
- Extraction Buffer
- Lysis Bufferl
- Sodium Acetate(pH5.2)
- 10mM Tris-HCl(pH8.0), 1mM EDTA
- 1mg/ml RNaseA
- E-coli culture
- TE(10mM Tris-HCl pH 8.0; 0.1mM EDTA)
- DW
<Process>
- Centrifuge 1.5ml of E-coli culture at 4οC for 5min, and then aspirate the supernatant.
- Add 0.3ml of Extraction Buffer to sample, and then vortex 1~2sec.
- Add 0.15mlof Lysis Buffer to solution, and then vortex 5~6sec.
- Incubate 15min at 50°C.
- Add 0.15ml of Sodium Acetate(pH5.2) to solution, and then vortex 1~2sec.
- Leave on ice for 15min.
- Centrifuge solution at 4οC for 15min, and then aspirate the supernatant.
- Add 0.3ml of H2O to the tube having the pellets, and then vortex thoroughly.
- Add 0.6ml of EtOH to solution, and then vortex thoroughly.
- Centrifuge solution for 10min, and then aspirate the supernatant.
- Add 70%EtOH to the tube having the pellets.
- Dry the pellets in the speed-vac for 5min, or air dry.
- Add TE Buffer to the dried pellets
Plasmid extraction from E-coli
<Materials>
- Binding Buffer (5M Guanidine Thiocyanate; 100mM Tris-HCl (pH7.0))
- Wash Buffer (10mM Tris-HCl (pH7.5))
- Silica gel solution
- TE (10mM Tris-HCl pH 8.0; 0.1mM EDTA)
- Sample
<Process>
- Centrifuge 1ml of the E-coli pre-culture on 12,000 rpm for 30sec, and then aspirate the supernatant.
- Add 100µl of the solution (50mM glucose; 10mM EDTA; 25ml Tris-HCl(pH 8.0)), and mix it gently
- Add 200µl of the 0.2N NaOH; 1% SDS, and mix it, and then incubate on ice for 5min.
- Add 150µl of 3M Potassium Acetate (pH 4.8)(5M Acetic acid; 3M Potassium), and mix it, and then incubate on ice for 5min.
- Centrifuge on 12,000 rpm for 5min.
- Add 400µl of Binding Buffer and 10µl of Silica gel solution to another tube and Vortex.
- Add the supernatant(in centrifuged tube) to solution of Binding Buffer and Silica gel, and Vortex.
- Centrifuge on 12,000 rpm for 5min, and then aspirate the supernatant.
- Add 800µl of Wash Buffer to tube and Vortex.
- Centrifuge on 12,000 rpm for 30sec, and then aspirate the supernatant.
- Add 800µl of Wash Buffer to tube and Vortex.
- Centrifuge on 12,000 rpm for 30sec, and then aspirate the supernatant.
- Dry the pellets in the speed-vac for 5min, or air dry.
- Add 50µl of TE Buffer to the dried pellets
Transformation with ECOS competent E-coli(NIPPON GENE)
<Materials>
- competent cell
- Plasmid solution
- LB plate(Amp)
<Process>
- Keep a competent cell(JM109) at RT and then on ice until melting.
- Add 5µl of the solution of ligated-recombinant plasmid DNA to the tube, which contains the competent cell, and mix it gently.
- Keep the tube on ice for 5min.
- Transfer the tube into 42°C water bath and keep it for 45sec.
- Vortex a suspension and spread it on a LB plate.
- Incubate the plate at 37°C for 12~16h.
Electrophoresis
<Materials>
- TAE Buffer 100ml
- Agarose 1.0g
- Marker 5.0µl
- Dye 1.0µl
- DNA solution
<Process>
- TAE Buffrer into agarose and Microwave for about 1min to dissolve the agarose.
- Pour the agarose slowly into the gel board.
- Set gel in a gel tank.
- Pour the TAE Buffer into the gel tank.
- Mix the marker, DW and Dye.
- Pour the mixture sample into well.
- Close the gel tank, switch on the power-source and run the gel at 100V for 20min.
- Switch off and unplug the gel tank and carry the gel to the machine to look at the progress of the gel.
<Marker>
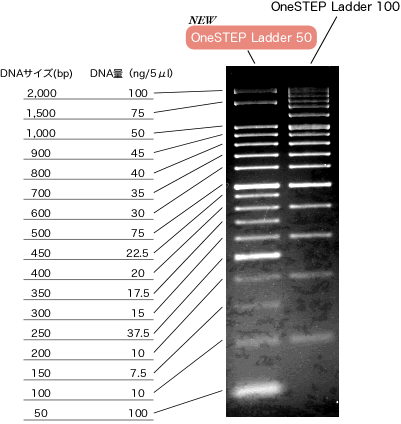
One of the marker we used is "OneSTEP Ladder 50"[http://www.nippongene.com/index/english/e_index.htm/ (NIPPONE GENE)]
DNA Digestion
<Material>
- DNA solution
- Digest enzyme
- ×10 Buffer
- DW
<Process>
- Add 5µl of DW and 2µl of ×10 Buffer into the tube, and mix it gently.
- Add 10µl of DNA solution and 1µl of Digest enzyme into the tube, and mix it gently.
- Incubate at 37°C for 5h.
Ligation with Ligation-convenience kit (NIPPON GENE)
<Materials>
- DNA solution
- 2×Ligation Mix (Ligation-convenience kit NIPPON GENE)
<Process>
- Prepare 10 μl of DNA solution to contain DNA fragment with appropriate mole ratio against vector DNA.
- Add 10μl of 2 × Ligation Mix to the DNA solution and mix well.
- Ligation reaction. Incubate 5-30min at 16°C.
- Apply DNA reaction mixture directly to transformation or in vitro packaging as it is.
DNA extraction with ISOPLANT(NIPPON GENE)
<Materials>
- Extraction Buffer 0.3ml
- Lysis Buffer 0.15ml
- Sodium Acetate(pH5.2) 0.15ml
- 10mM Tris-HCl(pH8.0), 1mM EDTA 0.1ml
- 1mg/ml RNaseA 1µl
- E-coli culture
- H20
- TE Buffer
<Process>
- Centrifuge 1.5ml of E-coli culture at 4οC for 5min, and then aspirate the supernatant.
- Add 0.3ml of Extraction Buffer to sample, and then vortex 1~2sec.
- Add 0.15mlof Lysis Buffer to solution, and then vortex 5~6sec.
- Incubate 15min at 50°C.
- Add 0.15ml of Sodium Acetate(pH5.2) to solution, and then vortex 1~2sec.
- Leave on ice for 15min
- Centrifuge solution at 4οC for 15min, and then aspirate the supernatant.
- Add 0.3ml of H2O to the tube having the pellets, and then vortex thoroughly.
- Add 0.6ml of EtOH to solution, and then vortex thoroughly.
- Centrifuge solution for 10min, and then aspirate the supernatant.
- Add 70%EtOH to the tube having the pellets.
- Dry the pellets in the speed-vac for 5min, or air dry.
- Add TE Buffer to the dried pellets
About Sample
| No. | template | fragment | Digest enzyme | bp |
| 1 | BBa_K208017 | Promoter-signal | AvrⅡ | 266 |
| 2 | E.coli genome | CyaA | NheⅠ | 2583 |
| 3 | BBa_l12521 | mRFP1-terminator | NheⅠ | 861 |
| 4 | BBa_K208017 | Promoter | AvrⅡ | 145 |
| 5 | BBa_K208017 | RBS-signal | SpeⅠ | 103 |
| 6 | BBa_l732901 | LacZ | SpeⅠ | 3093 |
| 7 | BBa_l12521 | terminator | NheⅠ | 167 |
| 8 | E. coli genome | CRP | AvrⅡ | 657 |
| 9 | BBa_K156010 | SBFP2 | XbaⅠ | 721 |
| 10 | BBa_J23110 | Promoter-RBS | SpeⅠ | 70 |
 "
"