Team:Alberta/Tour/biobytes
From 2010.igem.org
| (24 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
{{Team:Alberta/beginLeftSideBar|toc=NO|class=not-top}} | {{Team:Alberta/beginLeftSideBar|toc=NO|class=not-top}} | ||
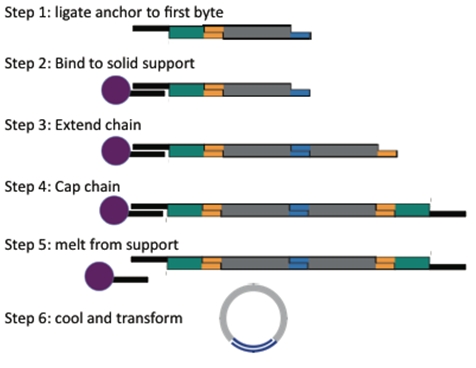
| + | [[Image:Team-alberta-biobyteprocess-tour.jpg|right|280px|thumb|This is the BioByte 2.0 system]] | ||
{{Team:Alberta/endLeftSideBar}} | {{Team:Alberta/endLeftSideBar}} | ||
| + | |||
{{Team:Alberta/beginRightSideBar|class=not-top}} | {{Team:Alberta/beginRightSideBar|class=not-top}} | ||
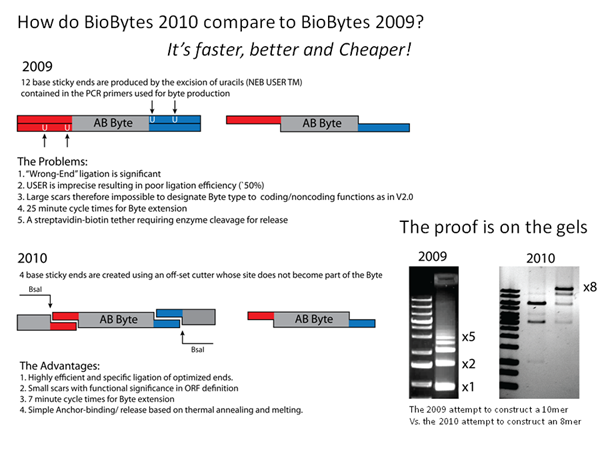
| + | [[Image:Alberta_BioByte2.0vs1.0.png|left|280px|thumb|GENOMIKON's BioByte 2.0 vs BioByte 1.0]] | ||
| + | <p></p> | ||
{{Team:Alberta/endRightSideBar}} | {{Team:Alberta/endRightSideBar}} | ||
{{Team:Alberta/beginMainContent|class=not-top}} | {{Team:Alberta/beginMainContent|class=not-top}} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ==Accelerated Construction== | |
| + | <div id="horiz-line"></div> | ||
| + | <p> | ||
| + | This year's project required a new approach to assembling DNA constructs. The need for a fast and efficient assembly standard was filled by BioBytes 2.0. BioBytes 2.0 is an innovative step forward from last year's BioBytes 1.0. | ||
| + | </p><p> | ||
| + | The new assembly method utilizes iron micro beads to anchor the DNA at a specified end. This allows for the DNA Bytes to be added in a controlled and sequential manner, ensuring the proper orientation and order of Bytes. This is made possible because of the design of our unique anchor part, which attaches to the iron micro bead at one end and to subsequent Bytes on the other. With the addition of our unique terminal cap Byte, the assembled construct will be able to circularize, and can then be transformed into ''E. coli''. | ||
| + | </p> | ||
{{Team:Alberta/endMainContent}} | {{Team:Alberta/endMainContent}} | ||
| - | |||
Latest revision as of 02:36, 28 October 2010
Accelerated Construction
This year's project required a new approach to assembling DNA constructs. The need for a fast and efficient assembly standard was filled by BioBytes 2.0. BioBytes 2.0 is an innovative step forward from last year's BioBytes 1.0.
The new assembly method utilizes iron micro beads to anchor the DNA at a specified end. This allows for the DNA Bytes to be added in a controlled and sequential manner, ensuring the proper orientation and order of Bytes. This is made possible because of the design of our unique anchor part, which attaches to the iron micro bead at one end and to subsequent Bytes on the other. With the addition of our unique terminal cap Byte, the assembled construct will be able to circularize, and can then be transformed into E. coli.
 "
"