Team:KIT-Kyoto/Project/Abstract
From 2010.igem.org
(→Abstract) |
(→Abstract) |
||
| (169 intermediate revisions not shown) | |||
| Line 60: | Line 60: | ||
<table border=0 width="965px" align="center"><tr><td> | <table border=0 width="965px" align="center"><tr><td> | ||
| - | <div aling="left">[[Team:KIT-Kyoto|Home]] > [[Team:KIT-Kyoto/Project/Abstract|Project]]</div></td><td><div align="right">Language : [[Team:KIT-Kyoto/Project/Abstract|English]] / [[Team:KIT-Kyoto/Project/AbstractJ|Japanese]]</div></td></tr></table> | + | <div aling="left">[[Team:KIT-Kyoto/Home|Home]] > [[Team:KIT-Kyoto/Project/Abstract|Project]]</div></td><td><div align="right">Language : [[Team:KIT-Kyoto/Project/Abstract|English]] / [[Team:KIT-Kyoto/Project/AbstractJ|Japanese]]</div></td></tr></table> |
<div id="NAKAMI"> | <div id="NAKAMI"> | ||
== Abstract == | == Abstract == | ||
| - | '''"''E.coli | + | ::'''"''E.coli'' Pen": Draw with your own color.''' |
| - | :: | + | :::Our team, KIT-Kyoto suggests an “''E.coli'' Pen” as a new Art Tool. This brand-new pen uses no ink but medium in which genetically modified ''E.coli'' has been cultured. The Pen is able to express more than four colors in various intensities with single bacterial culture. This will be achieved by constructing plasmids carrying genes coding for four different fluorescent proteins under the control of seven promoters having different sensitivity to oxidative stress. The ''E.coli'' carrying these plasmids will produce different colors with various intensities by differentially responding to the gradient of hydrogen peroxide treatment. Different from previous passive BioArt in iGEM, the genetically engineered “''E.coli'' Pen” provides an active and wonderful tool for us to purely enjoy the Art having a feeling for biotechnology. |
| + | <html><body><div align=center> | ||
| + | <IMG src="https://static.igem.org/mediawiki/2010/c/ca/20.jpg" onmouseover="this.src='https://static.igem.org/mediawiki/2010/6/62/11.jpg'" onmouseout="this.src='https://static.igem.org/mediawiki/2010/c/ca/20.jpg'"></div></body></html> | ||
== Introduction == | == Introduction == | ||
| - | :Our team, KIT-Kyoto has designed and carried out the project taking account of the key word "Make the science and technology more accessible to the public". <BR> | + | :::[[Image:88.jpg|right|200px]]Our team, KIT-Kyoto has designed and carried out the project taking account of the key word "Make the science and technology more accessible to the public". <BR> |
| - | :Recently, it is getting more and more serious problem that people think that the science is too difficult to understand. Moreover, it is expected that people are moving away from the science as the science becomes more and more complicated. Therefore, to overcome such a problem, we would like to offer more opportunities for the public to interact with the science especially for the person who is not interested in the modern science. <BR> | + | :::Recently, it is getting more and more serious problem that people think that the science is too difficult to understand. Moreover, it is expected that people are moving away from the science as the science becomes more and more complicated. Therefore, to overcome such a problem, we would like to offer more opportunities for the public to interact with the science especially for the person who is not interested in the modern science. <BR> |
| - | :In deciding our project, we thought it is the most important that anyone can readily enjoy the science and technology. We therefore decided to focus on "Bio art" in order to invent the artistic products by using biotechnology, since Art is more accessible to the public. Moreover, unifying the two apparently distant academic fields "Science" and "Art" is an educational philosophy of our University, Kyoto Institute of technology. | + | :::In deciding our project, we thought it is the most important that anyone can readily enjoy the science and technology. We therefore decided to focus on "Bio art" in order to invent the artistic products by using biotechnology, since Art is more accessible to the public. Moreover, unifying the two apparently distant academic fields "Science" and "Art" is an educational philosophy of our University, Kyoto Institute of technology. |
<html><body> | <html><body> | ||
<p style="width: 960px; margin:10px auto"> | <p style="width: 960px; margin:10px auto"> | ||
<ul class="acc" align="left"> | <ul class="acc" align="left"> | ||
| - | <li><a href="#2"> | + | <li><a href="#2"> >>Learn More BioArt</a> |
<ul class="fxmn" align=left> | <ul class="fxmn" align=left> | ||
<p> A brand-new form of art has emerged with the revolution of biology in the last ten years, this is Bio-Art. Adding new artistic elements, such as cells, DNA, genomes, proteins, enzymes, etc., this new form of artistic expression has as medium living matter, and the studios of the artists are biological laboratories, while high end technologies of genetic engineering, tissue culture and many others are their new painting brushes. | <p> A brand-new form of art has emerged with the revolution of biology in the last ten years, this is Bio-Art. Adding new artistic elements, such as cells, DNA, genomes, proteins, enzymes, etc., this new form of artistic expression has as medium living matter, and the studios of the artists are biological laboratories, while high end technologies of genetic engineering, tissue culture and many others are their new painting brushes. | ||
<p> BioArt is considered by most artists to be strictly limited to “living forms,” although there is some debate as to the stages at which matter can be considered to be alive or living. Creating living beings and practicing in the life sciences brings about ethical, social and aesthetic inquiry. | <p> BioArt is considered by most artists to be strictly limited to “living forms,” although there is some debate as to the stages at which matter can be considered to be alive or living. Creating living beings and practicing in the life sciences brings about ethical, social and aesthetic inquiry. | ||
| - | <p> Here, we aim at a completely new style of BioArt. In hitherto BioArt, the appreciator enjoys a work in the conventional way, that is he/she appreciates an artist’s resulting piece looking or touching it. The new style of BioArt we propose here is the enjoyment of making art itself. With the “<i> | + | <p> Here, we aim at a completely new style of BioArt. In hitherto BioArt, the appreciator enjoys a work in the conventional way, that is he/she appreciates an artist’s resulting piece looking or touching it. The new style of BioArt we propose here is the enjoyment of making art itself. With the “<i>E.coli</i> Pen” that we propose here the appreciator himself becomes part of the artistic work he is drawing enjoying the fascination of making the work and the work itself. |
</ul> | </ul> | ||
</li> | </li> | ||
</ul> | </ul> | ||
</html> | </html> | ||
| - | + | :::[[Image:bioart.jpg|thumb|left|200px|Already existing BioArt:wikipedia]]However, an already existing bio art does not meet the key word “Make the science and technology more accessible”, because that the preexisting bio art needs special equipment and the deep knowledge of biotechnology etc. We therefore decided to work on the development of a new art tool by which people could enjoy the bio art easily. <BR> | |
| - | :[[Image:bioart.jpg|thumb|left|200px| | + | :::Here we can propose a novel art tool, “''E.coli'' Pen” which draws paintings with innumerable colors as inks of the pen that are produced by a single <I>E. coli</I> bacteria. "<I>E.coli</I> Pen" would further develop the field of the bio art as a new art tool. Furthermore it is useful as an educational tool, since people can enjoy drawings with the heavenly color with happiness and at the same time they can study the molecular mechanism how the colors are made by genetic engineering.<br><br><br><br><br><br> |
| - | :Here we can propose a novel art tool, “''E.coli'' | + | ::We summarize how to make "''E.coli'' Pen" in the following section. |
| - | + | ::'''1. How to make an innumerable color in ''E. coli'' bacteria''' | |
| - | : | + | :::'''1-1. Use of light's three primary colors''' |
| - | :We summarize how to make "''E.coli'' | + | |
| - | :'''1. How to make an innumerable color in ''E. coli'' bacteria''' | + | |
| - | ::'''1-1. Use of light's three primary colors''' | + | |
:::Principle of light’s three primary colors (red, green, and blue) can be applied to create an innumerable color. The fluorescent protein can be used as a color source. Red Fluorescent Protein (RFP) can be used for Red, Green Fluorescent Protein (GFP) can be used for Green, and Cyan Fluorescent Protein (CFP) can be used for Blue. The color fluorescence is emitted when ultraviolet rays is irradiated. These fluorescent proteins are widely used in the field of the genetic engineering as the reporter gene. Therefore once we obtain the red, green, and blue fluorescent proteins in our hands, we can create an innumerable color by simply mixing these three fluorescent proteins with various ratios. At first we tried to produce the red, green, and blue fluorescent proteins in ''E. coli''. | :::Principle of light’s three primary colors (red, green, and blue) can be applied to create an innumerable color. The fluorescent protein can be used as a color source. Red Fluorescent Protein (RFP) can be used for Red, Green Fluorescent Protein (GFP) can be used for Green, and Cyan Fluorescent Protein (CFP) can be used for Blue. The color fluorescence is emitted when ultraviolet rays is irradiated. These fluorescent proteins are widely used in the field of the genetic engineering as the reporter gene. Therefore once we obtain the red, green, and blue fluorescent proteins in our hands, we can create an innumerable color by simply mixing these three fluorescent proteins with various ratios. At first we tried to produce the red, green, and blue fluorescent proteins in ''E. coli''. | ||
:::To achieve this, the multi-gene cassette shown in below was newly designed. | :::To achieve this, the multi-gene cassette shown in below was newly designed. | ||
| - | + | [[Image:gene.jpg|700px|center|]] | |
| - | :::In this multi-gene cassette, the genes encoding RFP (Red), GFP (Green) and CFP (Blue) are placed in a tandem array. Each gene can be controlled by different promoter that has different activity. In order to create different color, fluorescent proteins have to be produced in different ratios. This can be achieved by utilizing promoters having different activities. | + | :::In this multi-gene cassette, the genes encoding RFP (Red), GFP (Green) and CFP (Blue) are placed in a tandem array. Each gene can be controlled by different promoter that has different activity. In order to create different color, fluorescent proteins have to be produced in different ratios. This can be achieved by utilizing promoters having different activities.TM is a terminator. |
<BR> | <BR> | ||
::'''1-2 Selection of an appropriate promoter''' | ::'''1-2 Selection of an appropriate promoter''' | ||
| - | :::Then, what kind of promoter is actually appropriate for this purpose? The promoter activity has to be controlled by some regulating substances. The easily available substance that had not been used before in iGEM would be suitable for the purpose. If the substance is related to some human diseases or aging, the promoter responding to the substance may be used for diagnosis of the disease or aging in future, since the fluorescent proteins under the control of the promoter can be used as markers. From these standpoints we decided to use promoters responding to hydrogen peroxide. Hydrogen peroxide is one of the reactive oxygen species that oxidize nucleic acid, lipid and protein to result in aging of human cells and various diseases. Although effective use of oxygen is acquired during evolution, the defense system to the active oxygen generated by consumption of oxygen also has been acquired in the aerobe. There are a set of genes that are involved in this defense system. These genes can be induced by hydrogen peroxide. We have utilized the promoters of these genes to induce the fluorescent proteins. | + | :::[[Image:OXIDATIVESTRESS.GIF|right]]Then, what kind of promoter is actually appropriate for this purpose? The promoter activity has to be controlled by some regulating substances. The easily available substance that had not been used before in iGEM would be suitable for the purpose. If the substance is related to some human diseases or aging, the promoter responding to the substance may be used for diagnosis of the disease or aging in future, since the fluorescent proteins under the control of the promoter can be used as markers. From these standpoints we decided to use promoters responding to hydrogen peroxide. Hydrogen peroxide is one of the reactive oxygen species that oxidize nucleic acid, lipid and protein to result in aging of human cells and various diseases. Although effective use of oxygen is acquired during evolution, the defense system to the active oxygen generated by consumption of oxygen also has been acquired in the aerobe. There are a set of genes that are involved in this defense system. These genes can be induced by hydrogen peroxide. We have utilized the promoters of these genes to induce the fluorescent proteins. |
<BR> | <BR> | ||
| - | ::'''1-3 Mechanism of hydrogen peroxide response''' | + | ::'''1-3 Mechanism of hydrogen peroxide response''' |
:::''E. coli'' has two defense mechanisms against the active oxygen . One is OxyR response, and another is SoxRS response. | :::''E. coli'' has two defense mechanisms against the active oxygen . One is OxyR response, and another is SoxRS response. | ||
| - | :::In the OxyR response, OxyR works as a transcription factor. OxyR regulates activation or repression of the gene expression. Without active oxygen, OxyR is in an inactivated form. And with active oxygen, OxyR is activated and the activated OxyR can bind to the specific DNA sequence. OxyR binding | + | :::In the OxyR response, OxyR works as a transcription factor. OxyR regulates activation or repression of the gene expression. Without active oxygen, OxyR is in an inactivated form. And with active oxygen, OxyR is activated and the activated OxyR can bind to the specific DNA sequence. OxyR binding sites are found in promoters of a set of genes that respond to the active oxygen. In these active oxygen response genes, we focused on the promoter of ''ahpC'', ''dps'', ''oxyR'', ''sufA'', and ''yaiA''. These genes carry no recognition sequences of ''EcoR''Ⅰ, ''Xba''Ⅰ, ''Spe''Ⅰ,and ''Pst''Ⅰ restriction enzymes. These restriction enzyme sites would be used for cloning into the pSB1C3. The promoters of these genes cloned into pSB6A1 are listed in our Biobrick site. |
| - | :::In the SoxRS response, SoxR works as a transcription factor. SoxR is constantly expressed in cells. SoxR binds to the promoter of SoxS, and represses its transcription without hydrogen peroxide. With hydrogen peroxide, SoxR activates the SoxS gene transcription. SoxS also works as a transcription factor of two or more downstream genes responsible to the hydorogen peroxide. In these hydrogen peroxide response genes, we focused on the promoter of ''acrAB'' and ''sodA''. Both of these genes have no sequence of ''Eco''RⅠ, ''Xba''Ⅰ, ''Spe''Ⅰand ''Pst''Ⅰ restriction enzyme sites. These restriction enzyme sites would be used for cloning into the pSB1C3. The promoters of these two genes cloned into pSB6A1 are listed in our Biobrick site.[https://2010.igem.org/Team:KIT-Kyoto/Parts >>Go to the article of our Biobrick site. | + | :::In the SoxRS response, SoxR works as a transcription factor. SoxR is constantly expressed in cells. SoxR binds to the promoter of SoxS, and represses its transcription without hydrogen peroxide. With hydrogen peroxide, SoxR activates the SoxS gene transcription. SoxS also works as a transcription factor of two or more downstream genes responsible to the hydorogen peroxide. In these hydrogen peroxide response genes, we focused on the promoter of ''acrAB'' and ''sodA''. Both of these genes have no sequence of ''Eco''RⅠ, ''Xba''Ⅰ, ''Spe''Ⅰand ''Pst''Ⅰ restriction enzyme sites. These restriction enzyme sites would be used for cloning into the pSB1C3. The promoters of these two genes cloned into pSB6A1 are listed in our Biobrick site. |
| + | :::[https://2010.igem.org/Team:KIT-Kyoto/Parts >>Go to the article of our Biobrick site.] | ||
<BR> | <BR> | ||
::'''1-4 Active oxygen response promoters''' | ::'''1-4 Active oxygen response promoters''' | ||
| - | :::Out of these seven active oxygen response promoters, we here shows the structure of ahpC and sufA promoters. | + | :::Out of these seven active oxygen response promoters, we here shows the structure of ''ahpC'' and ''sufA'' promoters. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<BR> | <BR> | ||
| - | ::: | + | :::'''ahpC promoter'''<BR> |
| - | :[[Image: | + | :::[[Image:5.jpg|500px|left|thumb|Fig.1 OxyR is the transcriptional regulator for the expression of antioxidant genes in response to oxidative stress.The OxyR protein is constitutively produced and oxidized by hydrogen peroxide.The oxidized form(active form) of OxyR binds to the promoter region(OxyR binding region:OBR) of <I>ahpC</I> gene and promotes the transcription of the <I>ahpC</I> gene. ]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| - | ::: | + | |
| + | |||
| + | :::''ahpC-F'' are the genes involved in oxidative stress defense. Transcription of these genes is regulated in response to H<sub>2</sub>O<sub>2</sub>. The ''ahpC'' and ''ahpF'' genes encode the small subunit and large subunit of alkyl hydroperoxide reductase, respectively. The complex of AhpC and AhpF reduces alkyl hydroperoxide. The ''ahpC'' promoter is controlled by the OxyR protein, an oxidative stress responsive transcription factor. OxyR is constitutively expressed but activated only under oxidative stressed conditions caused by H<sub>2</sub>O<sub>2</sub>. The activated OxyR binds to the promoter region of the ''ahpC'' gene and promotes its transcription.<BR> | ||
<BR> | <BR> | ||
| - | :''' | + | :::'''sufA promoter'''<BR> |
| - | ::The '' | + | :::[[Image:81.jpg|left|600px|thumb|Fig.2 The <I>''sufA''</I> operon(<I>''sufABCDSE''</I>) is involved in the assembly of Fe-S clusters.Expression of the <I>''sufA''</I> gene is induced by superoxide generators and hydrogen peroxide.The <I>''sufA''</I>-proximal region also contains the OxyR binding region(OBR),and OxyR functions as one of the activators of the <I>''sufA''</I> operon.]]<br><br><br><br><br><br><br><br><br><br><br> |
| + | :::''sufA'' promoter is controlled by some transcription factors including OxyR, regulates the transcription of ''sufABCDSE'' operon encoding an alternative Fe-S cluster assembly system in ''E.coli''. We also constructed expression systems in which the ''sufA'' promoter controls the transcription of fluorescent protein genes and measured the fluorescent intensity. | ||
| + | <BR> | ||
| + | ::'''2. How is the pen that efficiently mixes the active oxygen and ''E. coli'' made?''' | ||
| + | :::The ''E.coli'' Pen changes the mixing rate for ''E. coli'' culture medium and hydrogen peroxide. The volume of each solution, placed in different tubes within the pen, can be regulated by the frequency and intensity of pressing the pen's piston which can be interchanged among both tubes. For example, to make red ink, one presses the pyston in the ''E. coli'' tube three times and then interchanges to the hydrogen peroxide tube and presses the piston twice. Or, to use green ink, press the'' E. coli'' tube five times and then the hydrogen peroxide tube only once. In this way, one is able to produce any color. This is a spectacular way of producing new colors that any conventional pen can not realize. The movie of the ''E.coli'' Pen is shown in the first page of wiki of our team. | ||
| + | <br> | ||
| + | :::[[Image:66.jpg|center|850px]] | ||
| + | <br> | ||
| + | :::[http://www.youtube.com/user/KITKyoto#p/u/9/DVVq0MWgRFk >>System of "<I>E.coli</I> Pen"] | ||
| + | :::[https://2010.igem.org/Team:KIT-Kyoto/DesignNote >>Go to the article of design note.] | ||
<BR> | <BR> | ||
== Materials & Methods == | == Materials & Methods == | ||
| - | <TABLE BORDER="0"><TR> | + | <TABLE BORDER="0"> |
| - | <TD>'''Materials'''</TD></TR> | + | <TR><TD>'''Materials'''</TD></TR> |
| - | <TR> | + | <TR><TD> '''Strains'''</TD><TR> |
| - | <TD> '''Strains'''</TD><TR> | + | <TR><TD> ''Esherichia coli''</TD><TD> DH5 Alpha</TD></TR> |
| - | <TR> | + | <TR><TD> '''Plasmids'''</TD></TR> |
| - | <TD> ''Esherichia coli''</TD><TD> DH5 Alpha</TD></TR> | + | <TR><TD> pSB3K3</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB3K3>>get more information]</TD></TR> |
| - | <TR> | + | <TR><TD> pSB6A1</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB6A1>>get more information]</TD></TR> |
| - | <TD> '''Plasmids'''</TD></TR> | + | <TR><TD> pSB1C3</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB1C3>>get more information]</TD></TR> |
| - | <TR> | + | <TR><TD> pSB1A2</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB1A2>>get more information]</TD></TR> |
| - | <TD> | + | <TR><TD> pSB1AK3</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB1AK3>>get more information]</TD></TR> |
| - | <TR> | + | <TR><TD> pSB4A5</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB4A5>>get more information]</TD></TR> |
| - | <TD> pSB6A1</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB6A1>>get more information]</TD></TR> | + | <TR><TD>'''Methods'''</TD></TR> |
| - | <TR> | + | <TR><TD COLSPAN="2">Our standard protocols are listed below. Please click each item for details.</TD></TR> |
| - | <TD> pSB1C3</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB1C3>>get more information]</TD></TR> | + | <TR><TD> '''List of protocols'''</TD></TR> |
| - | <TR> | + | <TR><TD> Bacterial transformation</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#tf >>Go protocol]</TD></TR> |
| - | <TD> pSB1A2</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB1A2>>get more information]</TD></TR> | + | <TR><TD> DNA miniprep</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#al >>Go protocol]</TD></TR> |
| - | <TR> | + | <TR><TD> PCR</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#pcr >>Go protocol]</TD></TR> |
| - | <TD> pSB1AK3</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB1AK3>>get more information]</TD></TR> | + | <TR><TD> DNA digestion by restriction enzymes</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#re >>Go protocol]</TD></TR> |
| - | <TR> | + | <TR><TD> Agarose gel electrophoresis</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#agar >>Go protocol]</TD></TR> |
| - | <TD> pSB4A5</TD><TD> [http://partsregistry.org/wiki/index.php/Part:pSB4A5>>get more information]</TD></TR> | + | <TR><TD> Isolation of DNA fragments from agarose gel</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#gel >>Go protocol]</TD></TR> |
| - | <TR> | + | <TR><TD> DNA ligation</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#lig >>Go protocol]</TD></TR> |
| - | <TD>'''Methods'''</TD></TR> | + | <TR><TD> '''Culture media'''</TD></TR> |
| - | <TR> | + | <TR><TD> LB culture medium</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#lb >>Go protocol]</TD></TR> |
| - | <TD COLSPAN="2">Our standard protocols are listed below. Please click each item for details.</TD></TR> | + | <TR><TD> 2xYT culture medium</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#yt >>Go protocol]</TD></TR> |
| - | <TR> | + | <TR><TD> SOB culture medium</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#sob >>Go protocol]</TD></TR> |
| - | <TD> '''List of protocols'''</TD></TR> | + | <TR><TD> SOC culture medium</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#soc >>Go protocol]</TD></TR> |
| - | <TR> | + | |
| - | <TD> Bacterial transformation</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#tf >>Go protocol]</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> DNA miniprep</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#al >>Go protocol]</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> PCR</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#pcr >>Go protocol]</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> DNA digestion by restriction enzymes</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#re >>Go protocol]</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> Agarose gel electrophoresis</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#agar >>Go protocol]</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> Isolation of DNA fragments from agarose gel</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#gel >>Go protocol]</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> DNA ligation</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#lig >>Go protocol]</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> '''Culture media'''</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> LB culture medium</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#lb >>Go protocol]</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> | + | |
| - | <TR> | + | |
| - | <TD> SOB culture medium</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#sob >>Go protocol]</TD></TR> | + | |
| - | <TR> | + | |
| - | <TD> SOC culture medium</TD><TD> [https://2010.igem.org/Team:KIT-Kyoto/Protocol#soc >>Go protocol]</TD></TR> | + | |
</TABLE> | </TABLE> | ||
| - | + | <br> | |
== Results & Discussion == | == Results & Discussion == | ||
| - | |||
=== 1. Susceptibility to H<sub>2</sub>O<sub>2</sub> in ''Escherichia coli'' === | === 1. Susceptibility to H<sub>2</sub>O<sub>2</sub> in ''Escherichia coli'' === | ||
| - | :In the ''E.coli'' Pen, we utilized the hydrogenperoxide (H<sub>2</sub>O<sub>2</sub>)-responsive system of ''E.coli'' cells to make different colors. It is well known that H<sub>2</sub>O<sub>2</sub> shows the concentration-dependent toxicity for ''E.coli'' cells. In order to determine the utilizable concentration of H<sub>2</sub>O<sub>2</sub> in the ''E.coli'' Pen, we monitored the susceptibility of ''E.coli'' cells ( | + | :::In the ''E.coli'' Pen, we utilized the hydrogenperoxide (H<sub>2</sub>O<sub>2</sub>)-responsive system of ''E.coli'' cells to make different colors. It is well known that H<sub>2</sub>O<sub>2</sub> shows the concentration-dependent toxicity for ''E.coli'' cells. In order to determine the utilizable concentration of H<sub>2</sub>O<sub>2</sub> in the ''E.coli'' Pen, we monitored the susceptibility of ''E.coli'' cells (DH5 Alpha) against H<sub>2</sub>O<sub>2</sub>. |
| - | '''<Experiment method>''' | + | :'''<Experiment method>''' |
| - | :'''The 1st day evening ''' | + | ::'''The 1st day evening ''' |
| - | ::Precultivation of | + | :::Precultivation of DH5 Alpha cells |
| - | :: | + | :::DH5 Alpha cells on a LB plate were picked up with the sterilized toothpick, inoculated into 2 ml of LB (amp-) medium in a test tube, and then precultured at 37˚C (overnight). |
| - | :'''The 2nd day''' | + | ::'''The 2nd day''' |
| - | ::Survival test | + | :::Survival test |
| - | ::All of the precultured | + | :::All of the precultured DH5 Alpha cells were transferred into 28 ml of a fresh LB (amp-) medium in a 100 ml Erlenmeyer flask. |
| - | ::↓ | + | :::↓ DH5 Alpha cells were cultivated at 37 ˚C until the optical density (OD600) of the medium became 0.4~0.6. |
| - | + | :::↓ The culture was equally dispensed (2 ml each) into sample tubes, and then cells were treated with various concentrations of H<sub>2</sub>O<sub>2</sub> (final concentrations were as follows: 1 pM, 10 pM, 200 pM. 1 nM, 10 nM, 100 nM, 1μM, 10μM, 100μM, and 1 mM) at 37 ˚C for 60 min. | |
| - | ::↓ | + | :::↓ Samples were diluted by mixing 1 ml of fresh LB medium and 1μl of each sample. |
| - | + | :::↓ 10 μl of the diluted sample was spread out on a LB plate (amp-) and incubated at 37 ˚C (overnight). | |
| - | ::↓ | + | |
| - | + | ||
| - | ::↓ | + | |
| - | + | ||
| - | :'''The 3rd day''' | + | ::'''The 3rd day''' |
| - | ::The number of colonies on the plate was counted and made a graph of the survival curve. | + | :::The number of colonies on the plate was counted and made a graph of the survival curve. |
| - | '''< | + | :'''<Results>'''<br> |
| - | :[[Image:0826-生存曲線.jpg]] | + | :::[[Image:0826-生存曲線.jpg|thumb|300px|left|Fig.3]]<br><br> |
| + | <br><br><br><br><br><br><br><br><br><br><br> | ||
| + | :::Representative data from five independent experiments are shown. | ||
| - | : | + | :'''<Discussion>''' |
| + | :::As shown in the graph, 1mM of H<sub>2</sub>O<sub>2</sub> was the lethal concentration for ''E.coli'' cells. On the other hand, less than 0.000001mM(=1nM) H<sub>2</sub>O<sub>2</sub> has no influence on the survival of ''E.coli'' cells. These results suggest that a range of 1nM-1mM of H<sub>2</sub>O<sub>2</sub> is the utilizable concentration for the ''E.coli'' Pen. | ||
| - | '''< | + | :'''<References>''' |
| - | : | + | :::Application serial number in Japan (P2004-551020) "Ozone generation goes through an antibody or neutrophile" by Novartis Akuchiengezerushafuto and the Scrips Res. Inst. |
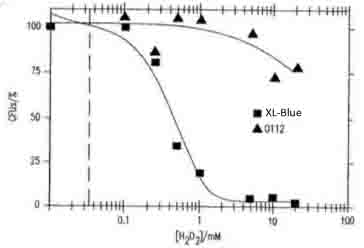
| - | + | :::[[Image:0826-引用画像.jpg|left|300px|thumb|Fig.4]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br> | |
| - | : | + | :::Fig.4 shows the concentration of H<sub>2</sub>O<sub>2</sub> dependent toxicity for the viability of ''E.coli'' XL1- blue and O112a, c serotype. Our data is supported by this reference. |
| - | + | <BR> | |
| - | :[[Image:0826-引用画像.jpg]] | + | |
| - | : | + | |
=== 2. Compare the performances of the high copy vector and low copy vector === | === 2. Compare the performances of the high copy vector and low copy vector === | ||
| - | :In making the E.coli Pen, we thought that it was important to select “ink.” The ''E.coli'' Pen, which focused on writing or drawing, is a first attempt in iGEM, so we thought that we should start to select an appropriate vector to produce “ink”, fluorescent proteins. As “ink”, it is necessary to keep writing or drawing clearly over the long term, and we therefore selected an expression vector with high transcriptional efficiency. | + | :::In making the <I>E.coli</I> Pen, we thought that it was important to select “ink.” The ''E.coli'' Pen, which focused on writing or drawing, is a first attempt in iGEM, so we thought that we should start to select an appropriate vector to produce “ink”, fluorescent proteins. As “ink”, it is necessary to keep writing or drawing clearly over the long term, and we therefore selected an expression vector with high transcriptional efficiency. |
| - | :In general, pSB6 is a low copy plasmid which makes it more suitable for protein production than the high copy vector. However this information cannot be found in any sites related to iGEM. Consequently, we tried to compare the performances of the high copy vector pSB1 and low copy vector pSB6. | + | :::In general, pSB6 is a low copy plasmid which makes it more suitable for protein production than the high copy vector. However this information cannot be found in any sites related to iGEM. Consequently, we tried to compare the performances of the high copy vector pSB1 and low copy vector pSB6. |
| - | + | ||
| + | |||
| + | :'''<Experiment method>''' | ||
| + | :::''E.coli'' cells were cultivated at 37 ˚C until the optical density (O.D.600) of the medium became 0.4~0.5. Cells were suspended in PBS to obtain the initial O.D.600 = 0.5 because the LB medium has auto fluorescence, then the fluorescent intensity was measured using Thermo Labosystems Floreskan Ascent CF. | ||
| + | |||
| + | :'''<Results>''' | ||
| + | :::<table border=1> | ||
| + | ::<tr><td> </td><td colspan=3 align=center>Vector</td></tr> | ||
| + | ::<tr><td align=center>Fluorescent intensity</td><td align=center>pSB1A2(promoter less GFP)</td><td align=center>pSB1A2(High Copy Vector)</td><td align=center>pSB6A1(Low Copy Vector)</td></tr> | ||
| + | ::<tr><td align=center>1</td><td align=right>0.145</td><td align=right>0.349</td><td align=right>4.33</td></tr> | ||
| + | ::<tr><td align=center>2</td><td align=right>0.146</td><td align=right>0.324</td><td align=right>4.34</td></tr> | ||
| + | ::<tr><td align=center>3</td><td align=right>0.146</td><td align=right>0.328</td><td align=right>4.42</td></tr> | ||
| + | ::<tr><td align=center>Ave</td><td align=right>0.146</td><td align=right>0.337</td><td align=right>4.36</td></tr></table> | ||
| + | <br> | ||
| + | :::[[Image:g1.jpg|left|400px|thumb|Fig.5]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | :'''<Discussion>''' | ||
| + | :::Clearly the Low copy vector has much higher transcriptional efficiency of [GFP] than the High copy vector. | ||
| + | :::So we selected a Low copy vector (pSB6A1) to produce fluorescent proteins. | ||
| + | <BR> | ||
=== 3. Constructing an expression system === | === 3. Constructing an expression system === | ||
| - | :We constructed an expression system in which the | + | :We constructed an expression system in which the active oxygen response promoter controls the transcription of fluorescent protein genes on pSB6, and transformed this vector into DH5 Alpha competent cells. Here we show an expressing system by using ''ahpC'' promoter and ''sufA'' promoter among 7 promoters which we constructed. We then examined the fluorescent intensity using the cells treated with H<sub>2</sub>O<sub>2</sub> as follows: |
| - | |||
| - | |||
| - | ''' | + | :'''<Experiment method>''' |
| + | ::''E.coli'' cells in the log-phase were treated with H<sub>2</sub>O<sub>2</sub> at 37℃. The fluorescent intensity was measured every 10 minutes until 80minutes. After the treatment with H<sub>2</sub>O<sub>2</sub>, cells were suspended in PBS to obtain the initial OD600 = 0.5 because the LB medium has auto fluorescence, then the fluorescent intensity was measured using Thermo Labosystems Floreskan Ascent CF. | ||
| - | |||
| - | + | :'''<Results>'''<BR> | |
| - | + | ::'''3-1. ''ahpC'' (BBa_K362001) ''' | |
| - | '''< | + | |
| - | : | + | |
| - | + | :::[[Image:AhpC.jpg|left|400px|thumb|Fig.6]] | |
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| - | '''< | + | :::Fig.6 indicates that the ''ahpC'' promoter responded to H<sub>2</sub>O<sub>2</sub> in a dose-dependent manner and its activity increased with time. Especially, the treatment with 10 μM to 1mM H<sub>2</sub>O<sub>2</sub> for 40 to 70 min increased the fluorescent intensity remarkably. After 70 min, the intensity decayed with time. |
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| + | ::'''3-2. ''sufA'' (BBa_K362005) ''' | ||
| + | |||
| + | :::We also constructed expression systems in which the ''sufA'' promoter controls the transcription of fluorescent protein genes and measured the fluorescent intensity. | ||
| + | |||
| + | :::[[Image:SufA graph.jpg|left|400px|thumb|Fig.7]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | |||
| + | :::Fig.7 shows that the ''sufA'' promoter responded to H<sub>2</sub>O<sub>2</sub> stress in a dose-dependent manner.The ''sufA'' promoter region is located upstream of the ''sufA'' operon and SufA protein acts as an assembly scaffold. Since levels of SufA protein are maximized quite rapidly, the activity of the ''sufA'' promoter was increased and sustained for at least 80 min. We think that the concentration-dependent activation of the ''sufA'' promoter also reflects the levels of SufA protein.<BR> | ||
| + | |||
| + | |||
| + | :'''<Discussion>''' | ||
| + | ::Our results seem quite reasonable and suggest a possibility that we can adjust the expression levels of fluorescent genes by H<sub>2</sub>O<sub>2</sub> concentration and by time. Therefore, we expect that lots of colors can be created using this system. Our expression system must be a powerful tool for the construction of the ''E.coli'' Pen. | ||
<BR> | <BR> | ||
| Line 258: | Line 262: | ||
=== 4. Making Inks === | === 4. Making Inks === | ||
| - | :This section consists of two parts. The first one is about making inks with various colors. The other is the actual art works drawn using the inks we made. | + | ::This section consists of two parts. The first one is about making inks with various colors. The other is the actual art works drawn using the inks we made. |
| - | '''4-1. Making inks with various colors.''' | + | :'''4-1. Making inks with various colors.''' |
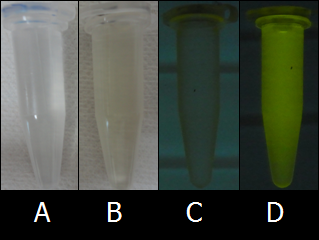
| - | :[[Image:KITfig1.png|thumb|left|319px|Fig. | + | ::[[Image:KITfig1.png|thumb|left|319px|Fig.8 Autofluorescence of LB medium and PBS.(A) PBS without UV light, (B) LB medium without UV light, (C) PBS under UV light, (D) LB medium under UV light.]] |
<BR><BR> | <BR><BR> | ||
| - | |||
| - | |||
| + | ::Although we initially used the LB medium as a solvent, the colors of inks were not so clear because LB medium has high levels of background fluorescence (autofluorescence). To find the proper solvent for BioArt, we tried out various solutions. Fig.8 shows the comparison of autofluorescence between PBS buffer and LB medium. As shown in Figs. 8C and 8D, PBS buffer has lower levels of autofluorescence than LB medium. As far as we tried out, PBS buffer was one of the best solvent of ''E.coli'' cells and we adopted PBS buffer as the solvent for the ''E.coli'' Pen. | ||
| - | We succeeded to make several ink colors such as Red, Orange, Yellow, and Green. Additionally, we succeeded to develop various colors by mixing the Green ink and Red ink. | + | |
| + | ::We succeeded to make several ink colors such as Red ink, Orange ink, Yellow ink, and Green ink. Additionally, we succeeded to develop various colors by mixing the Green ink and Red ink. | ||
<BR><BR><BR><BR><BR><BR> | <BR><BR><BR><BR><BR><BR> | ||
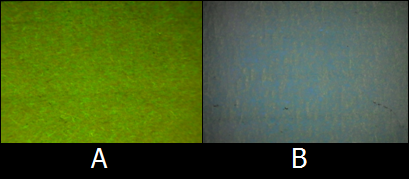
| - | :[[Image:KITfig2.png|thumb|left|220px|Fig. | + | :::[[Image:KITfig2.png|thumb|left|220px|Fig.9 Yellow is made by mixing Red ink and Green ink.(A)Red ink by RFP expression under UV light, (B) Green ink by GFP expression under UV light, (C) Yellow ink by mixing RFP:GFP=1:2 under UV light.]]<BR><BR><BR> |
| - | <BR> | + | |
| - | + | ||
| - | To make Green and Red, we used K362001 on pSB6A1, a GFP expression vector, and J04450 on pSB1C3, RFP expression vector, respectively. | + | ::To make Green ink and Red ink, we used K362001 on pSB6A1, a GFP expression vector, and J04450 on pSB1C3, RFP expression vector, respectively. |
| - | Furthermore, we mixed Green ink and Red ink to make other colors such as Orange and Yellow. Fig. | + | Furthermore, we mixed Green ink and Red ink to make other colors such as Orange ink and Yellow ink. Fig.9C shows one of a new color ink, Yellow, made by mixing equal amounts of Green ink and Red ink. |
<BR><BR> | <BR><BR> | ||
<BR><BR><BR><BR><BR> | <BR><BR><BR><BR><BR> | ||
| - | + | <BR> | |
| - | <BR><BR><BR><BR> | + | <BR><BR><BR> |
| - | + | <BR> | |
| - | :[[Image:KITfig3.png|thumb|left|400px|Fig. | + | :::[[Image:KITfig3.png|thumb|left|400px|Fig.10 Various colors of our bio inks.(A)Red ink, (B) Orange ink, (C) Yellow ink ,(D)GFP ink, and (E)PBS under UV light.]] |
<BR><BR> | <BR><BR> | ||
<BR> | <BR> | ||
<BR><BR> | <BR><BR> | ||
| - | To make Orange, we mixed Green ink and Red ink in a ratio of 1 : 2. Of course, we can create various colors by mixing inks in a various ratio. | + | ::To make Orange ink, we mixed Green ink and Red ink in a ratio of 1 : 2. To make Yellow ink, we mixed Green ink and Red ink in a ratio of 2 : 1. Of course, we can create various colors by mixing inks in a various ratio. |
| Line 295: | Line 298: | ||
| - | '''4-2. Art works with bio inks.''' | + | ::'''4-2. Art works with bio inks.''' |
| - | :At first, we used newspaper as drawing papers. However, we found that newspaper has high levels of intrinsic fluorescence and is not suitable for BioArt (Fig. | + | :::At first, we used newspaper as drawing papers. However, we found that newspaper has high levels of intrinsic fluorescence and is not suitable for BioArt (Fig.11). Indeed, pictures drawn with bio inks on newspaper were dull and not clear. Therefore, we tried to find the proper paper for BioArt. As shown in Fig.11, ‘‘Kimwipes’’, a type of cleaning tissue commonly used in laboratories, did not show almost any intrinsic fluorescence. Why do some papers have intrinsic fluorescence? It is well known that lignin, commonly derived from wood, has a strong intrinsic fluorescence, and some papers are rich in lignin. That is one of reasons why some papers showed high levels of intrinsic fluorescence. On the other hand, little lignin is contained in Kimwipes. Therefore, we concluded that Kimwipes is the best drawing papers for BioArt with the ''E.coli'' Pen. |
| - | :[[Image:KITfig5.png|thumb|left|409px|Fig. | + | :::[[Image:KITfig5.png|thumb|left|409px|Fig.11 Intrinsic fluorescence of newspaper and Kimwipes. Newspaper (A) and Kimwipes (B) under UV light.]]<BR><BR> |
<BR> | <BR> | ||
<BR><BR> | <BR><BR> | ||
| - | Using bio inks and Kimwipes, we drew and painted lots pictures, marks, letters, etc. Various examples of BioArt works are shown as follows. Please enjoy our art works !!! | + | :::Using bio inks and Kimwipes, we drew and painted lots pictures, marks, letters, etc. Various examples of BioArt works are shown as follows. Please enjoy our art works !!! |
<BR><BR><BR><BR> | <BR><BR><BR><BR> | ||
<BR><BR><BR> | <BR><BR><BR> | ||
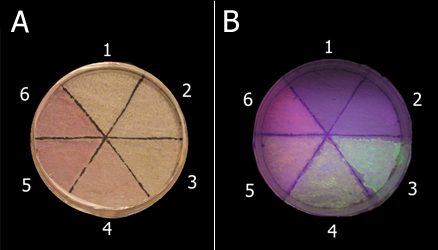
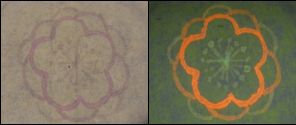
| - | :[[Image:KITfig6.png|thumb|left|438px|Fig. | + | :::[[Image:KITfig6.png|thumb|left|438px|Fig.12 |
Samples are as follows: 1, no ink; 2, PBS; 3, Green ink; 4, Yellow ink; 5, Orange ink; 6, Red ink. Samples were irradiated without UV light, (A) or with UV light (B). ]]<BR><BR> | Samples are as follows: 1, no ink; 2, PBS; 3, Green ink; 4, Yellow ink; 5, Orange ink; 6, Red ink. Samples were irradiated without UV light, (A) or with UV light (B). ]]<BR><BR> | ||
<BR><BR><BR> | <BR><BR><BR> | ||
| - | :We used these inks in order to make samples. Next figures shows some picture that we drew. | + | :::We used these inks in order to make samples. Next figures shows some picture that we drew. |
<BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | <BR><BR><BR><BR><BR><BR><BR><BR><BR><BR> | ||
<table><tr><td> | <table><tr><td> | ||
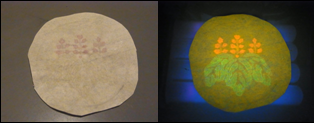
| - | <div align="center">[[Image:KITfig7.png|left|thumb|274px|Fig. | + | <div align="center">[[Image:KITfig7.png|left|thumb|274px|Fig.13 Maple leaves]] |
| - | [[Image:KITfig8.png|thumb|left|178px|Fig. | + | :::[[Image:KITfig8.png|thumb|left|178px|Fig.14 Bamboo]]</div> |
</td></tr><tr><td> | </td></tr><tr><td> | ||
<div align="center"> | <div align="center"> | ||
| - | [[Image:KITfig9.png|thumb|left|314px|Fig. | + | [[Image:KITfig9.png|thumb|left|314px|Fig.15 Emblem of Samurai ,Toyotomi Hideyosi]] |
| - | [[Image:KITfig10.png|thumb|left|296px|Fig. | + | [[Image:KITfig10.png|thumb|left|296px|Fig.16 Japanese plum]] |
</div> | </div> | ||
</td></tr><tr><td> | </td></tr><tr><td> | ||
<div align="center"> | <div align="center"> | ||
| - | [[Image:KITfig11.png|thumb|left|273px|Fig. | + | [[Image:KITfig11.png|thumb|left|273px|Fig.17 Cherry blossoms]] |
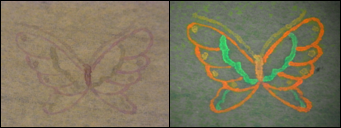
| - | [[Image:KITfig12.png|thumb|left|341px|Fig. | + | [[Image:KITfig12.png|thumb|left|341px|Fig.18 Butterfly]] |
</div> | </div> | ||
</td></tr><tr><td> | </td></tr><tr><td> | ||
<div align="center"> | <div align="center"> | ||
| - | [[Image:KITfig13.png|thumb|left|236px|Fig. | + | [[Image:KITfig13.png|thumb|left|236px|Fig.19 Ukiyoe]] |
| - | [[Image:KITfig14.png|thumb|left|151px|Fig. | + | [[Image:KITfig14.png|thumb|left|151px|Fig.20 Carp]] |
| - | [[Image:KITfig16.png|thumb|left|154px|Fig. | + | [[Image:KITfig16.png|thumb|left|154px|Fig.21 Crane ]] |
| - | [[Image:KITfig17.png|thumb|left|169px|Fig. | + | [[Image:KITfig17.png|thumb|left|169px|Fig.22 Ginkgo]] |
</div> | </div> | ||
</td></tr></table> | </td></tr></table> | ||
| Line 345: | Line 348: | ||
== References == | == References == | ||
| - | # [http://www.ncbi.nlm.nih.gov/pubmed/11443092 PMID: 11443092] Ming Zheng, Xunde Wang, Bernard Doan, Karen A. Lewis, Thomas D. Schneider, Gisela Storz,Computation-Directed Identification of OxyR DNA Binding Sites in <I>Escherichia coli</I>,Journal of Bacteriology,2001 Aug;183(15):4571-79. | + | :# [http://www.ncbi.nlm.nih.gov/pubmed/11443092 PMID: 11443092] Ming Zheng, Xunde Wang, Bernard Doan, Karen A. Lewis, Thomas D. Schneider, Gisela Storz,Computation-Directed Identification of OxyR DNA Binding Sites in <I>Escherichia coli</I>,Journal of Bacteriology,2001 Aug;183(15):4571-79. |
| - | # [http://www.ncbi.nlm.nih.gov/pubmed/10913087 PMID: 10913087] Urs A. Ochsner, Michel L. Vasil, Eyad Alsabbagh, Kislay Parvatiyar, Daniel Hassett,Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF.,Journal of Bacteriology,2000 Aug;182(16):4533-44. | + | :# [http://www.ncbi.nlm.nih.gov/pubmed/10913087 PMID: 10913087] Urs A. Ochsner, Michel L. Vasil, Eyad Alsabbagh, Kislay Parvatiyar, Daniel Hassett,Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF.,Journal of Bacteriology,2000 Aug;182(16):4533-44. |
| - | # [http://ci.nii.ac.jp/naid/110001710476 NAID: 110001710476] Nunoshiba Tatsuo,Mechanisms for the oxidative stress response and oxidative mutagenesis in <I>Escherichia coli</I>.,Environmental mutagen research,2001 June 30;23(1):23-32. | + | :# [http://ci.nii.ac.jp/naid/110001710476 NAID: 110001710476] Nunoshiba Tatsuo,Mechanisms for the oxidative stress response and oxidative mutagenesis in <I>Escherichia coli</I>.,Environmental mutagen research,2001 June 30;23(1):23-32. |
| - | # [http://www.ncbi.nlm.nih.gov/pubmed/12876288 PMID: 12876288] Laurent Loiseau, Sandrine Ollagnier-de-Choudens, Laurence Nachin, Mare Fontecave, Frederie Barras,Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase.,The Journal of Biological Chemistry,2003 Oct 3;278(40):38352-9. | + | :# [http://www.ncbi.nlm.nih.gov/pubmed/12876288 PMID: 12876288] Laurent Loiseau, Sandrine Ollagnier-de-Choudens, Laurence Nachin, Mare Fontecave, Frederie Barras,Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase.,The Journal of Biological Chemistry,2003 Oct 3;278(40):38352-9. |
| - | # [http://www.ncbi.nlm.nih.gov/pubmed/18849427 PMID: 18849427] Joon-Hee Lee, Won-Sik Yeo, Jung-Hye Roe,Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor.,Molecular Microbiology,2008 Dec;190(24):8244-7. | + | :# [http://www.ncbi.nlm.nih.gov/pubmed/18849427 PMID: 18849427] Joon-Hee Lee, Won-Sik Yeo, Jung-Hye Roe,Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor.,Molecular Microbiology,2008 Dec;190(24):8244-7. |
| - | # [http://www.ncbi.nlm.nih.gov/pubmed/16621810 PMID: 16621810] Ramakrishnan Balasubramanian, Gaozhong Shen, Donald A. Bryant, John H. Golbeck,Regulatory Roles for IscA and SufA in Iron Homeostasis and Redox Stress Responses in the Cyanobacterium Synechococcus sp. Strain PCC 7002.,Journal of Bacteriology,2006 May;188(9):3182-91. | + | :# [http://www.ncbi.nlm.nih.gov/pubmed/16621810 PMID: 16621810] Ramakrishnan Balasubramanian, Gaozhong Shen, Donald A. Bryant, John H. Golbeck,Regulatory Roles for IscA and SufA in Iron Homeostasis and Redox Stress Responses in the Cyanobacterium Synechococcus sp. Strain PCC 7002.,Journal of Bacteriology,2006 May;188(9):3182-91. |
| - | # [http://www.ncbi.nlm.nih.gov/pubmed/11443091 PMID: 11443091] Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G.,DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide.,Journal of Bacteriology,2001 Aug;183(15):4562-70. | + | :# [http://www.ncbi.nlm.nih.gov/pubmed/11443091 PMID: 11443091] Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G.,DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide.,Journal of Bacteriology,2001 Aug;183(15):4562-70. |
| - | # [http://www.ncbi.nlm.nih.gov/pubmed/12889026 PMID: 12889026] Lu C, Bentley WE, Rao G.,Comparisons of oxidative stress response genes in aerobic <I>Escherichia coli</I> fermentations.,Biotechnology and Bioengineering,2003 Sep 30;83(7):864-70. | + | :# [http://www.ncbi.nlm.nih.gov/pubmed/12889026 PMID: 12889026] Lu C, Bentley WE, Rao G.,Comparisons of oxidative stress response genes in aerobic <I>Escherichia coli</I> fermentations.,Biotechnology and Bioengineering,2003 Sep 30;83(7):864-70. |
| - | # [http://ci.nii.ac.jp/naid/110000056227 NAID: 110000056227] Takebe So, Azuma Megumi, Yoshida Megumi, Yoshida Aya,Effect of cAMP-CRP regulation on sodA promoter activity.,Kyoto Women's University food journal,1995 Dec 10;50:37-42. | + | :# [http://ci.nii.ac.jp/naid/110000056227 NAID: 110000056227] Takebe So, Azuma Megumi, Yoshida Megumi, Yoshida Aya,Effect of cAMP-CRP regulation on sodA promoter activity.,Kyoto Women's University food journal,1995 Dec 10;50:37-42. |
| - | # [http://www.ncbi.nlm.nih.gov/pubmed/10585871 PMID: 10585871] Mitsumoto A, Kim KR, Oshima G, Kunimoto M, Okawa K, Iwamatsu A, Nakagawa Y.,Glyoxalase I is a novel nitric-oxide-responsive protein.,The Biochemical journal,1999 Dec 15;344 Pt 3:837-44. | + | :# [http://www.ncbi.nlm.nih.gov/pubmed/10585871 PMID: 10585871] Mitsumoto A, Kim KR, Oshima G, Kunimoto M, Okawa K, Iwamatsu A, Nakagawa Y.,Glyoxalase I is a novel nitric-oxide-responsive protein.,The Biochemical journal,1999 Dec 15;344 Pt 3:837-44. |
| - | # [http://www.ekouhou.net/%E6%8A%97%E4%BD%93%E3%81%BE%E3%81%9F%E3%81%AF%E5%A5%BD%E4%B8%AD%E7%90%83%E3%82%92%E4%BB%8B%E3%81%99%E3%82%8B%E3%82%AA%E3%82%BE%E3%83%B3%E7%94%9F%E6%88%90/disp-A,2006-506613.html PCT/EP2003/012710] Novartis, The Scripps Research Institute,Generation of ozone through antibody or neutrophil,2004 May 27. | + | :# [http://www.ekouhou.net/%E6%8A%97%E4%BD%93%E3%81%BE%E3%81%9F%E3%81%AF%E5%A5%BD%E4%B8%AD%E7%90%83%E3%82%92%E4%BB%8B%E3%81%99%E3%82%8B%E3%82%AA%E3%82%BE%E3%83%B3%E7%94%9F%E6%88%90/disp-A,2006-506613.html PCT/EP2003/012710] Novartis, The Scripps Research Institute,Generation of ozone through antibody or neutrophil,2004 May 27. |
{{Template:KIT-Kyoto-1}} | {{Template:KIT-Kyoto-1}} | ||
Latest revision as of 21:17, 27 October 2010
 
|
|
Home > Project | Language : English / Japanese |
Abstract
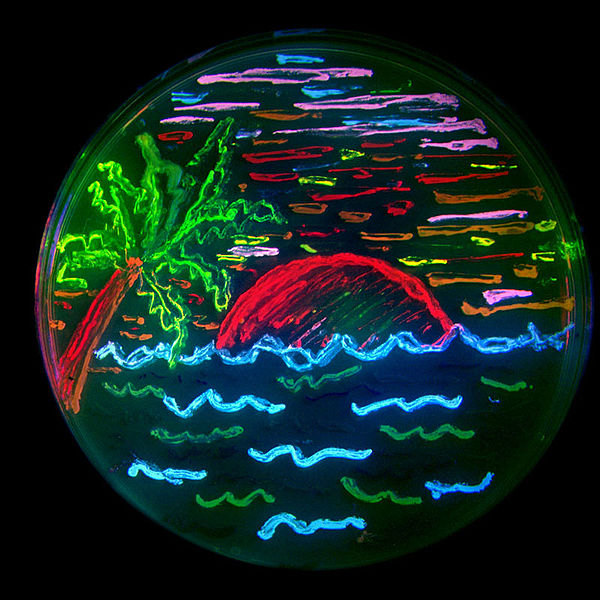
- "E.coli Pen": Draw with your own color.
- Our team, KIT-Kyoto suggests an “E.coli Pen” as a new Art Tool. This brand-new pen uses no ink but medium in which genetically modified E.coli has been cultured. The Pen is able to express more than four colors in various intensities with single bacterial culture. This will be achieved by constructing plasmids carrying genes coding for four different fluorescent proteins under the control of seven promoters having different sensitivity to oxidative stress. The E.coli carrying these plasmids will produce different colors with various intensities by differentially responding to the gradient of hydrogen peroxide treatment. Different from previous passive BioArt in iGEM, the genetically engineered “E.coli Pen” provides an active and wonderful tool for us to purely enjoy the Art having a feeling for biotechnology.
- "E.coli Pen": Draw with your own color.

Introduction
- Our team, KIT-Kyoto has designed and carried out the project taking account of the key word "Make the science and technology more accessible to the public".
- Recently, it is getting more and more serious problem that people think that the science is too difficult to understand. Moreover, it is expected that people are moving away from the science as the science becomes more and more complicated. Therefore, to overcome such a problem, we would like to offer more opportunities for the public to interact with the science especially for the person who is not interested in the modern science.
- In deciding our project, we thought it is the most important that anyone can readily enjoy the science and technology. We therefore decided to focus on "Bio art" in order to invent the artistic products by using biotechnology, since Art is more accessible to the public. Moreover, unifying the two apparently distant academic fields "Science" and "Art" is an educational philosophy of our University, Kyoto Institute of technology.
- Our team, KIT-Kyoto has designed and carried out the project taking account of the key word "Make the science and technology more accessible to the public".
- >>Learn More BioArt
A brand-new form of art has emerged with the revolution of biology in the last ten years, this is Bio-Art. Adding new artistic elements, such as cells, DNA, genomes, proteins, enzymes, etc., this new form of artistic expression has as medium living matter, and the studios of the artists are biological laboratories, while high end technologies of genetic engineering, tissue culture and many others are their new painting brushes.
BioArt is considered by most artists to be strictly limited to “living forms,” although there is some debate as to the stages at which matter can be considered to be alive or living. Creating living beings and practicing in the life sciences brings about ethical, social and aesthetic inquiry.
Here, we aim at a completely new style of BioArt. In hitherto BioArt, the appreciator enjoys a work in the conventional way, that is he/she appreciates an artist’s resulting piece looking or touching it. The new style of BioArt we propose here is the enjoyment of making art itself. With the “E.coli Pen” that we propose here the appreciator himself becomes part of the artistic work he is drawing enjoying the fascination of making the work and the work itself.
- However, an already existing bio art does not meet the key word “Make the science and technology more accessible”, because that the preexisting bio art needs special equipment and the deep knowledge of biotechnology etc. We therefore decided to work on the development of a new art tool by which people could enjoy the bio art easily.
- Here we can propose a novel art tool, “E.coli Pen” which draws paintings with innumerable colors as inks of the pen that are produced by a single E. coli bacteria. "E.coli Pen" would further develop the field of the bio art as a new art tool. Furthermore it is useful as an educational tool, since people can enjoy drawings with the heavenly color with happiness and at the same time they can study the molecular mechanism how the colors are made by genetic engineering.
- However, an already existing bio art does not meet the key word “Make the science and technology more accessible”, because that the preexisting bio art needs special equipment and the deep knowledge of biotechnology etc. We therefore decided to work on the development of a new art tool by which people could enjoy the bio art easily.
- We summarize how to make "E.coli Pen" in the following section.
- 1. How to make an innumerable color in E. coli bacteria
- 1-1. Use of light's three primary colors
- Principle of light’s three primary colors (red, green, and blue) can be applied to create an innumerable color. The fluorescent protein can be used as a color source. Red Fluorescent Protein (RFP) can be used for Red, Green Fluorescent Protein (GFP) can be used for Green, and Cyan Fluorescent Protein (CFP) can be used for Blue. The color fluorescence is emitted when ultraviolet rays is irradiated. These fluorescent proteins are widely used in the field of the genetic engineering as the reporter gene. Therefore once we obtain the red, green, and blue fluorescent proteins in our hands, we can create an innumerable color by simply mixing these three fluorescent proteins with various ratios. At first we tried to produce the red, green, and blue fluorescent proteins in E. coli.
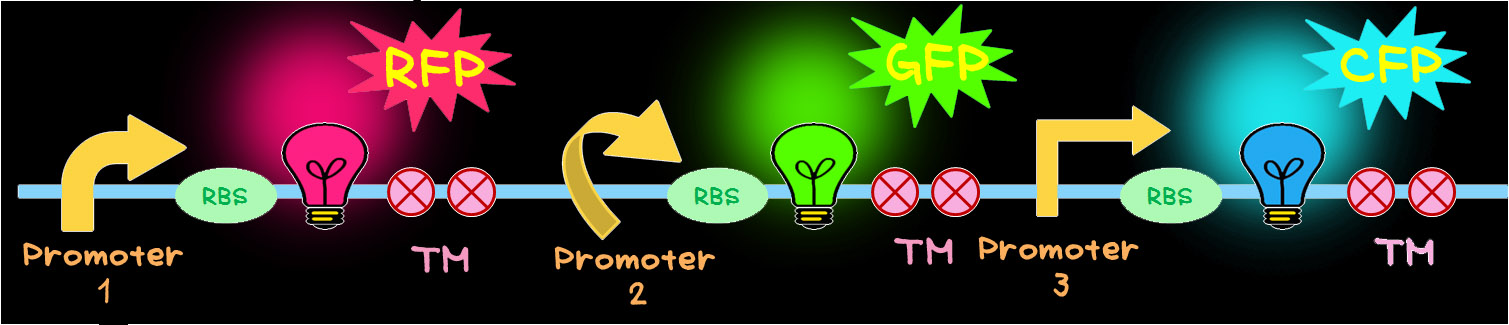
- To achieve this, the multi-gene cassette shown in below was newly designed.
- In this multi-gene cassette, the genes encoding RFP (Red), GFP (Green) and CFP (Blue) are placed in a tandem array. Each gene can be controlled by different promoter that has different activity. In order to create different color, fluorescent proteins have to be produced in different ratios. This can be achieved by utilizing promoters having different activities.TM is a terminator.
- 1-2 Selection of an appropriate promoter
- Then, what kind of promoter is actually appropriate for this purpose? The promoter activity has to be controlled by some regulating substances. The easily available substance that had not been used before in iGEM would be suitable for the purpose. If the substance is related to some human diseases or aging, the promoter responding to the substance may be used for diagnosis of the disease or aging in future, since the fluorescent proteins under the control of the promoter can be used as markers. From these standpoints we decided to use promoters responding to hydrogen peroxide. Hydrogen peroxide is one of the reactive oxygen species that oxidize nucleic acid, lipid and protein to result in aging of human cells and various diseases. Although effective use of oxygen is acquired during evolution, the defense system to the active oxygen generated by consumption of oxygen also has been acquired in the aerobe. There are a set of genes that are involved in this defense system. These genes can be induced by hydrogen peroxide. We have utilized the promoters of these genes to induce the fluorescent proteins.
- 1-2 Selection of an appropriate promoter
- 1-3 Mechanism of hydrogen peroxide response
- E. coli has two defense mechanisms against the active oxygen . One is OxyR response, and another is SoxRS response.
- In the OxyR response, OxyR works as a transcription factor. OxyR regulates activation or repression of the gene expression. Without active oxygen, OxyR is in an inactivated form. And with active oxygen, OxyR is activated and the activated OxyR can bind to the specific DNA sequence. OxyR binding sites are found in promoters of a set of genes that respond to the active oxygen. In these active oxygen response genes, we focused on the promoter of ahpC, dps, oxyR, sufA, and yaiA. These genes carry no recognition sequences of EcoRⅠ, XbaⅠ, SpeⅠ,and PstⅠ restriction enzymes. These restriction enzyme sites would be used for cloning into the pSB1C3. The promoters of these genes cloned into pSB6A1 are listed in our Biobrick site.
- In the SoxRS response, SoxR works as a transcription factor. SoxR is constantly expressed in cells. SoxR binds to the promoter of SoxS, and represses its transcription without hydrogen peroxide. With hydrogen peroxide, SoxR activates the SoxS gene transcription. SoxS also works as a transcription factor of two or more downstream genes responsible to the hydorogen peroxide. In these hydrogen peroxide response genes, we focused on the promoter of acrAB and sodA. Both of these genes have no sequence of EcoRⅠ, XbaⅠ, SpeⅠand PstⅠ restriction enzyme sites. These restriction enzyme sites would be used for cloning into the pSB1C3. The promoters of these two genes cloned into pSB6A1 are listed in our Biobrick site.
- >>Go to the article of our Biobrick site.
- 1-3 Mechanism of hydrogen peroxide response
- 1-4 Active oxygen response promoters
- Out of these seven active oxygen response promoters, we here shows the structure of ahpC and sufA promoters.
- 1-4 Active oxygen response promoters
- ahpC promoter
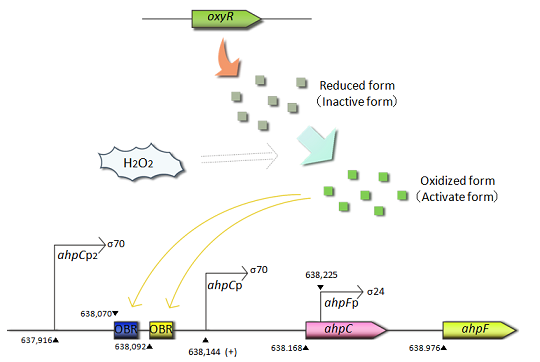 Fig.1 OxyR is the transcriptional regulator for the expression of antioxidant genes in response to oxidative stress.The OxyR protein is constitutively produced and oxidized by hydrogen peroxide.The oxidized form(active form) of OxyR binds to the promoter region(OxyR binding region:OBR) of ahpC gene and promotes the transcription of the ahpC gene.
Fig.1 OxyR is the transcriptional regulator for the expression of antioxidant genes in response to oxidative stress.The OxyR protein is constitutively produced and oxidized by hydrogen peroxide.The oxidized form(active form) of OxyR binds to the promoter region(OxyR binding region:OBR) of ahpC gene and promotes the transcription of the ahpC gene.
- ahpC promoter
- ahpC-F are the genes involved in oxidative stress defense. Transcription of these genes is regulated in response to H2O2. The ahpC and ahpF genes encode the small subunit and large subunit of alkyl hydroperoxide reductase, respectively. The complex of AhpC and AhpF reduces alkyl hydroperoxide. The ahpC promoter is controlled by the OxyR protein, an oxidative stress responsive transcription factor. OxyR is constitutively expressed but activated only under oxidative stressed conditions caused by H2O2. The activated OxyR binds to the promoter region of the ahpC gene and promotes its transcription.
- ahpC-F are the genes involved in oxidative stress defense. Transcription of these genes is regulated in response to H2O2. The ahpC and ahpF genes encode the small subunit and large subunit of alkyl hydroperoxide reductase, respectively. The complex of AhpC and AhpF reduces alkyl hydroperoxide. The ahpC promoter is controlled by the OxyR protein, an oxidative stress responsive transcription factor. OxyR is constitutively expressed but activated only under oxidative stressed conditions caused by H2O2. The activated OxyR binds to the promoter region of the ahpC gene and promotes its transcription.
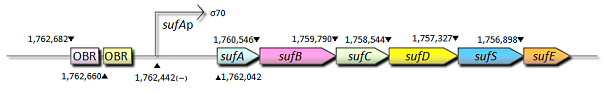
- sufA promoter
- sufA promoter
- sufA promoter is controlled by some transcription factors including OxyR, regulates the transcription of sufABCDSE operon encoding an alternative Fe-S cluster assembly system in E.coli. We also constructed expression systems in which the sufA promoter controls the transcription of fluorescent protein genes and measured the fluorescent intensity.
- 2. How is the pen that efficiently mixes the active oxygen and E. coli made?
- The E.coli Pen changes the mixing rate for E. coli culture medium and hydrogen peroxide. The volume of each solution, placed in different tubes within the pen, can be regulated by the frequency and intensity of pressing the pen's piston which can be interchanged among both tubes. For example, to make red ink, one presses the pyston in the E. coli tube three times and then interchanges to the hydrogen peroxide tube and presses the piston twice. Or, to use green ink, press the E. coli tube five times and then the hydrogen peroxide tube only once. In this way, one is able to produce any color. This is a spectacular way of producing new colors that any conventional pen can not realize. The movie of the E.coli Pen is shown in the first page of wiki of our team.
- 2. How is the pen that efficiently mixes the active oxygen and E. coli made?
- [http://www.youtube.com/user/KITKyoto#p/u/9/DVVq0MWgRFk >>System of "E.coli Pen"]
- >>Go to the article of design note.
Materials & Methods
| Materials | |
| Strains | |
| Esherichia coli | DH5 Alpha |
| Plasmids | |
| pSB3K3 | [http://partsregistry.org/wiki/index.php/Part:pSB3K3>>get more information] |
| pSB6A1 | [http://partsregistry.org/wiki/index.php/Part:pSB6A1>>get more information] |
| pSB1C3 | [http://partsregistry.org/wiki/index.php/Part:pSB1C3>>get more information] |
| pSB1A2 | [http://partsregistry.org/wiki/index.php/Part:pSB1A2>>get more information] |
| pSB1AK3 | [http://partsregistry.org/wiki/index.php/Part:pSB1AK3>>get more information] |
| pSB4A5 | [http://partsregistry.org/wiki/index.php/Part:pSB4A5>>get more information] |
| Methods | |
| Our standard protocols are listed below. Please click each item for details. | |
| List of protocols | |
| Bacterial transformation | >>Go protocol |
| DNA miniprep | >>Go protocol |
| PCR | >>Go protocol |
| DNA digestion by restriction enzymes | >>Go protocol |
| Agarose gel electrophoresis | >>Go protocol |
| Isolation of DNA fragments from agarose gel | >>Go protocol |
| DNA ligation | >>Go protocol |
| Culture media | |
| LB culture medium | >>Go protocol |
| 2xYT culture medium | >>Go protocol |
| SOB culture medium | >>Go protocol |
| SOC culture medium | >>Go protocol |
Results & Discussion
1. Susceptibility to H2O2 in Escherichia coli
- In the E.coli Pen, we utilized the hydrogenperoxide (H2O2)-responsive system of E.coli cells to make different colors. It is well known that H2O2 shows the concentration-dependent toxicity for E.coli cells. In order to determine the utilizable concentration of H2O2 in the E.coli Pen, we monitored the susceptibility of E.coli cells (DH5 Alpha) against H2O2.
- <Experiment method>
- The 1st day evening
- Precultivation of DH5 Alpha cells
- DH5 Alpha cells on a LB plate were picked up with the sterilized toothpick, inoculated into 2 ml of LB (amp-) medium in a test tube, and then precultured at 37˚C (overnight).
- The 2nd day
- Survival test
- All of the precultured DH5 Alpha cells were transferred into 28 ml of a fresh LB (amp-) medium in a 100 ml Erlenmeyer flask.
- ↓ DH5 Alpha cells were cultivated at 37 ˚C until the optical density (OD600) of the medium became 0.4~0.6.
- ↓ The culture was equally dispensed (2 ml each) into sample tubes, and then cells were treated with various concentrations of H2O2 (final concentrations were as follows: 1 pM, 10 pM, 200 pM. 1 nM, 10 nM, 100 nM, 1μM, 10μM, 100μM, and 1 mM) at 37 ˚C for 60 min.
- ↓ Samples were diluted by mixing 1 ml of fresh LB medium and 1μl of each sample.
- ↓ 10 μl of the diluted sample was spread out on a LB plate (amp-) and incubated at 37 ˚C (overnight).
- The 3rd day
- The number of colonies on the plate was counted and made a graph of the survival curve.
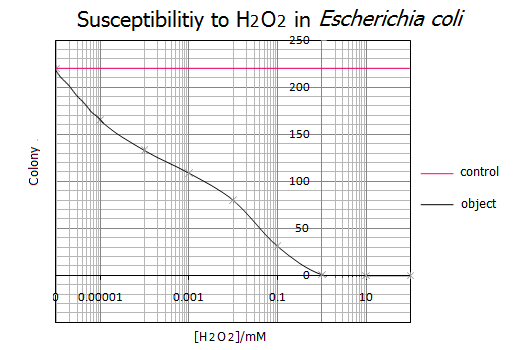
- <Results>
- Representative data from five independent experiments are shown.
- <Discussion>
- As shown in the graph, 1mM of H2O2 was the lethal concentration for E.coli cells. On the other hand, less than 0.000001mM(=1nM) H2O2 has no influence on the survival of E.coli cells. These results suggest that a range of 1nM-1mM of H2O2 is the utilizable concentration for the E.coli Pen.
- <References>
- Application serial number in Japan (P2004-551020) "Ozone generation goes through an antibody or neutrophile" by Novartis Akuchiengezerushafuto and the Scrips Res. Inst.
- Fig.4 shows the concentration of H2O2 dependent toxicity for the viability of E.coli XL1- blue and O112a, c serotype. Our data is supported by this reference.
2. Compare the performances of the high copy vector and low copy vector
- In making the E.coli Pen, we thought that it was important to select “ink.” The E.coli Pen, which focused on writing or drawing, is a first attempt in iGEM, so we thought that we should start to select an appropriate vector to produce “ink”, fluorescent proteins. As “ink”, it is necessary to keep writing or drawing clearly over the long term, and we therefore selected an expression vector with high transcriptional efficiency.
- In general, pSB6 is a low copy plasmid which makes it more suitable for protein production than the high copy vector. However this information cannot be found in any sites related to iGEM. Consequently, we tried to compare the performances of the high copy vector pSB1 and low copy vector pSB6.
- <Experiment method>
- E.coli cells were cultivated at 37 ˚C until the optical density (O.D.600) of the medium became 0.4~0.5. Cells were suspended in PBS to obtain the initial O.D.600 = 0.5 because the LB medium has auto fluorescence, then the fluorescent intensity was measured using Thermo Labosystems Floreskan Ascent CF.
- <Results>
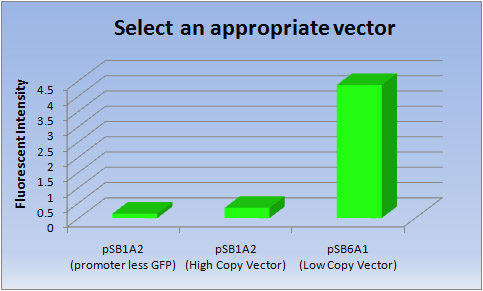
Vector Fluorescent intensity pSB1A2(promoter less GFP) pSB1A2(High Copy Vector) pSB6A1(Low Copy Vector) 1 0.145 0.349 4.33 2 0.146 0.324 4.34 3 0.146 0.328 4.42 Ave 0.146 0.337 4.36
- <Discussion>
- Clearly the Low copy vector has much higher transcriptional efficiency of [GFP] than the High copy vector.
- So we selected a Low copy vector (pSB6A1) to produce fluorescent proteins.
3. Constructing an expression system
- We constructed an expression system in which the active oxygen response promoter controls the transcription of fluorescent protein genes on pSB6, and transformed this vector into DH5 Alpha competent cells. Here we show an expressing system by using ahpC promoter and sufA promoter among 7 promoters which we constructed. We then examined the fluorescent intensity using the cells treated with H2O2 as follows:
- <Experiment method>
- E.coli cells in the log-phase were treated with H2O2 at 37℃. The fluorescent intensity was measured every 10 minutes until 80minutes. After the treatment with H2O2, cells were suspended in PBS to obtain the initial OD600 = 0.5 because the LB medium has auto fluorescence, then the fluorescent intensity was measured using Thermo Labosystems Floreskan Ascent CF.
- <Results>
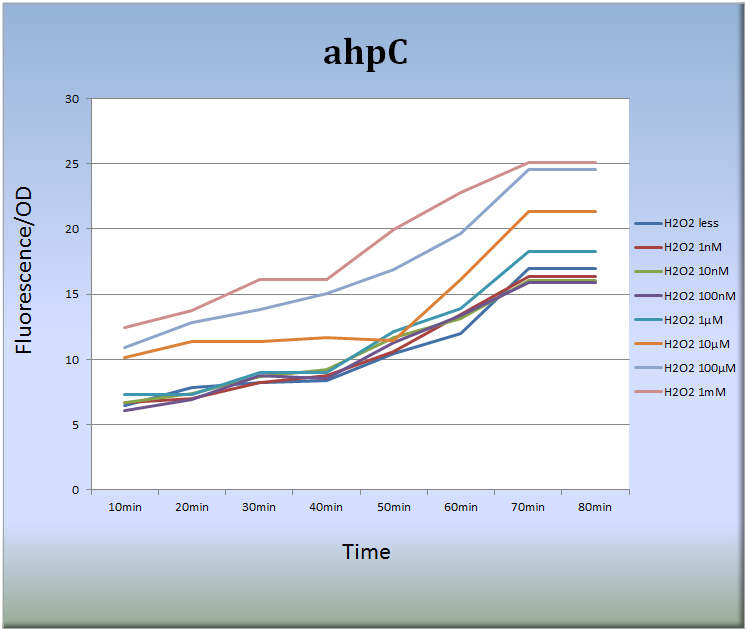
- 3-1. ahpC (BBa_K362001)
- Fig.6 indicates that the ahpC promoter responded to H2O2 in a dose-dependent manner and its activity increased with time. Especially, the treatment with 10 μM to 1mM H2O2 for 40 to 70 min increased the fluorescent intensity remarkably. After 70 min, the intensity decayed with time.
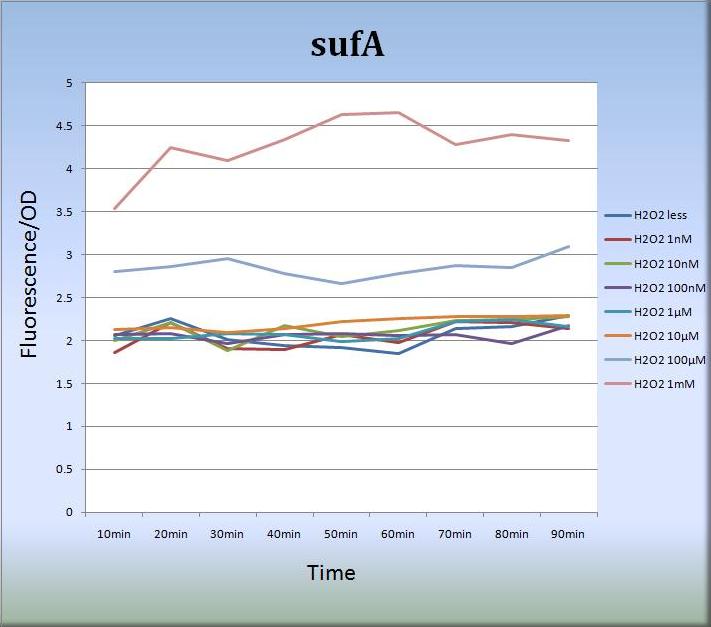
- 3-2. sufA (BBa_K362005)
- We also constructed expression systems in which the sufA promoter controls the transcription of fluorescent protein genes and measured the fluorescent intensity.
- Fig.7 shows that the sufA promoter responded to H2O2 stress in a dose-dependent manner.The sufA promoter region is located upstream of the sufA operon and SufA protein acts as an assembly scaffold. Since levels of SufA protein are maximized quite rapidly, the activity of the sufA promoter was increased and sustained for at least 80 min. We think that the concentration-dependent activation of the sufA promoter also reflects the levels of SufA protein.
- Fig.7 shows that the sufA promoter responded to H2O2 stress in a dose-dependent manner.The sufA promoter region is located upstream of the sufA operon and SufA protein acts as an assembly scaffold. Since levels of SufA protein are maximized quite rapidly, the activity of the sufA promoter was increased and sustained for at least 80 min. We think that the concentration-dependent activation of the sufA promoter also reflects the levels of SufA protein.
- <Discussion>
- Our results seem quite reasonable and suggest a possibility that we can adjust the expression levels of fluorescent genes by H2O2 concentration and by time. Therefore, we expect that lots of colors can be created using this system. Our expression system must be a powerful tool for the construction of the E.coli Pen.
4. Making Inks
- This section consists of two parts. The first one is about making inks with various colors. The other is the actual art works drawn using the inks we made.
- 4-1. Making inks with various colors.
- Although we initially used the LB medium as a solvent, the colors of inks were not so clear because LB medium has high levels of background fluorescence (autofluorescence). To find the proper solvent for BioArt, we tried out various solutions. Fig.8 shows the comparison of autofluorescence between PBS buffer and LB medium. As shown in Figs. 8C and 8D, PBS buffer has lower levels of autofluorescence than LB medium. As far as we tried out, PBS buffer was one of the best solvent of E.coli cells and we adopted PBS buffer as the solvent for the E.coli Pen.
- We succeeded to make several ink colors such as Red ink, Orange ink, Yellow ink, and Green ink. Additionally, we succeeded to develop various colors by mixing the Green ink and Red ink.
- To make Green ink and Red ink, we used K362001 on pSB6A1, a GFP expression vector, and J04450 on pSB1C3, RFP expression vector, respectively.
Furthermore, we mixed Green ink and Red ink to make other colors such as Orange ink and Yellow ink. Fig.9C shows one of a new color ink, Yellow, made by mixing equal amounts of Green ink and Red ink.
- To make Orange ink, we mixed Green ink and Red ink in a ratio of 1 : 2. To make Yellow ink, we mixed Green ink and Red ink in a ratio of 2 : 1. Of course, we can create various colors by mixing inks in a various ratio.
- 4-2. Art works with bio inks.
- At first, we used newspaper as drawing papers. However, we found that newspaper has high levels of intrinsic fluorescence and is not suitable for BioArt (Fig.11). Indeed, pictures drawn with bio inks on newspaper were dull and not clear. Therefore, we tried to find the proper paper for BioArt. As shown in Fig.11, ‘‘Kimwipes’’, a type of cleaning tissue commonly used in laboratories, did not show almost any intrinsic fluorescence. Why do some papers have intrinsic fluorescence? It is well known that lignin, commonly derived from wood, has a strong intrinsic fluorescence, and some papers are rich in lignin. That is one of reasons why some papers showed high levels of intrinsic fluorescence. On the other hand, little lignin is contained in Kimwipes. Therefore, we concluded that Kimwipes is the best drawing papers for BioArt with the E.coli Pen.
- Using bio inks and Kimwipes, we drew and painted lots pictures, marks, letters, etc. Various examples of BioArt works are shown as follows. Please enjoy our art works !!!
- We used these inks in order to make samples. Next figures shows some picture that we drew.
References
- [http://www.ncbi.nlm.nih.gov/pubmed/11443092 PMID: 11443092] Ming Zheng, Xunde Wang, Bernard Doan, Karen A. Lewis, Thomas D. Schneider, Gisela Storz,Computation-Directed Identification of OxyR DNA Binding Sites in Escherichia coli,Journal of Bacteriology,2001 Aug;183(15):4571-79.
- [http://www.ncbi.nlm.nih.gov/pubmed/10913087 PMID: 10913087] Urs A. Ochsner, Michel L. Vasil, Eyad Alsabbagh, Kislay Parvatiyar, Daniel Hassett,Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF.,Journal of Bacteriology,2000 Aug;182(16):4533-44.
- [http://ci.nii.ac.jp/naid/110001710476 NAID: 110001710476] Nunoshiba Tatsuo,Mechanisms for the oxidative stress response and oxidative mutagenesis in Escherichia coli.,Environmental mutagen research,2001 June 30;23(1):23-32.
- [http://www.ncbi.nlm.nih.gov/pubmed/12876288 PMID: 12876288] Laurent Loiseau, Sandrine Ollagnier-de-Choudens, Laurence Nachin, Mare Fontecave, Frederie Barras,Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase.,The Journal of Biological Chemistry,2003 Oct 3;278(40):38352-9.
- [http://www.ncbi.nlm.nih.gov/pubmed/18849427 PMID: 18849427] Joon-Hee Lee, Won-Sik Yeo, Jung-Hye Roe,Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor.,Molecular Microbiology,2008 Dec;190(24):8244-7.
- [http://www.ncbi.nlm.nih.gov/pubmed/16621810 PMID: 16621810] Ramakrishnan Balasubramanian, Gaozhong Shen, Donald A. Bryant, John H. Golbeck,Regulatory Roles for IscA and SufA in Iron Homeostasis and Redox Stress Responses in the Cyanobacterium Synechococcus sp. Strain PCC 7002.,Journal of Bacteriology,2006 May;188(9):3182-91.
- [http://www.ncbi.nlm.nih.gov/pubmed/11443091 PMID: 11443091] Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G.,DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide.,Journal of Bacteriology,2001 Aug;183(15):4562-70.
- [http://www.ncbi.nlm.nih.gov/pubmed/12889026 PMID: 12889026] Lu C, Bentley WE, Rao G.,Comparisons of oxidative stress response genes in aerobic Escherichia coli fermentations.,Biotechnology and Bioengineering,2003 Sep 30;83(7):864-70.
- [http://ci.nii.ac.jp/naid/110000056227 NAID: 110000056227] Takebe So, Azuma Megumi, Yoshida Megumi, Yoshida Aya,Effect of cAMP-CRP regulation on sodA promoter activity.,Kyoto Women's University food journal,1995 Dec 10;50:37-42.
- [http://www.ncbi.nlm.nih.gov/pubmed/10585871 PMID: 10585871] Mitsumoto A, Kim KR, Oshima G, Kunimoto M, Okawa K, Iwamatsu A, Nakagawa Y.,Glyoxalase I is a novel nitric-oxide-responsive protein.,The Biochemical journal,1999 Dec 15;344 Pt 3:837-44.
- [http://www.ekouhou.net/%E6%8A%97%E4%BD%93%E3%81%BE%E3%81%9F%E3%81%AF%E5%A5%BD%E4%B8%AD%E7%90%83%E3%82%92%E4%BB%8B%E3%81%99%E3%82%8B%E3%82%AA%E3%82%BE%E3%83%B3%E7%94%9F%E6%88%90/disp-A,2006-506613.html PCT/EP2003/012710] Novartis, The Scripps Research Institute,Generation of ozone through antibody or neutrophil,2004 May 27.
 "
"