Team:Lethbridge/Notebook/Lab Work/August
From 2010.igem.org
(→Aug 11, 2010) |
Liszabruder (Talk | contribs) |
||
| (71 intermediate revisions not shown) | |||
| Line 209: | Line 209: | ||
<b>Method:</b> Used [[Team:Lethbridge/Notebook/Protocols|Restriction of Plasmid DNA]] protocol. | <b>Method:</b> Used [[Team:Lethbridge/Notebook/Protocols|Restriction of Plasmid DNA]] protocol. | ||
| - | * A front | + | * A front vector was made made in sRBS ,mRBS, dT plasmids using EcoRI and Xbal enzymes |
* pDNA that was cut out of plasmid for a front vector was pBAD, TetR, dT and pLacI using EcoRI and SpeI enzymes | * pDNA that was cut out of plasmid for a front vector was pBAD, TetR, dT and pLacI using EcoRI and SpeI enzymes | ||
* A back vector was made in sRBS and mRBS plasmids with PstI and SpeI | * A back vector was made in sRBS and mRBS plasmids with PstI and SpeI | ||
| Line 252: | Line 252: | ||
</table> | </table> | ||
*Added 48.8 µL of Master Mix to each PCR reaction<br><br><br> | *Added 48.8 µL of Master Mix to each PCR reaction<br><br><br> | ||
| + | |||
| + | |||
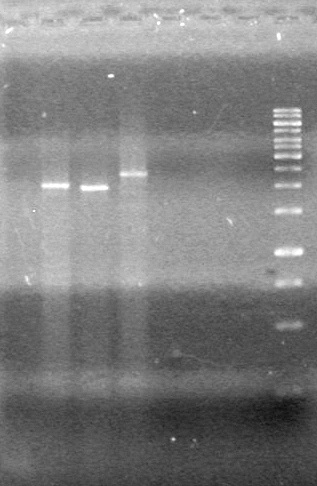
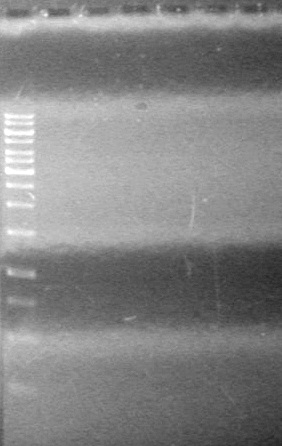
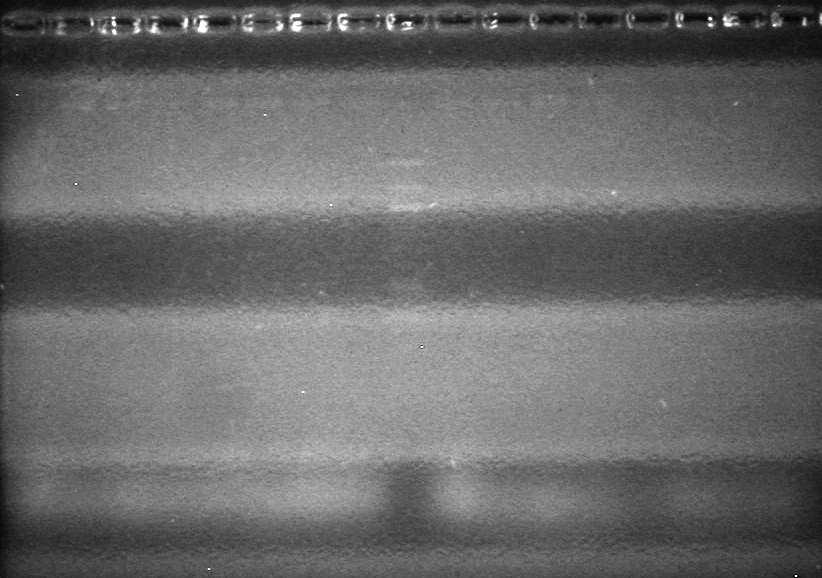
<b>Objective:</b> Complete maxipreps of Lumazine (K249002), EYFP (E0030), and ECFP (E0020) and run on 1% agarose gel.<br> | <b>Objective:</b> Complete maxipreps of Lumazine (K249002), EYFP (E0030), and ECFP (E0020) and run on 1% agarose gel.<br> | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 262: | Line 264: | ||
</table> | </table> | ||
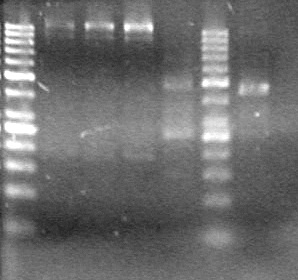
*Ran at 100V for 42 minutes. Stained in EtBr for 15 minutes.<br> | *Ran at 100V for 42 minutes. Stained in EtBr for 15 minutes.<br> | ||
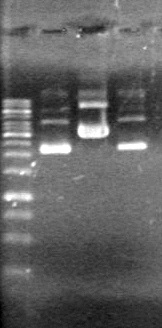
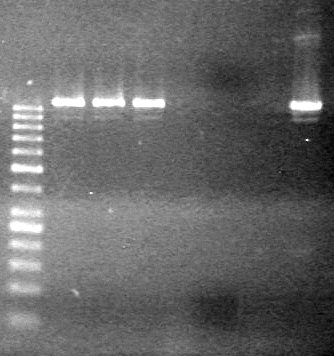
| - | <b>Results:</b> | + | <b>Results:</b> [[image:Lethbridge_100803JV Maxiprep.JPG|50px]]<br><br><br> |
| + | |||
| + | |||
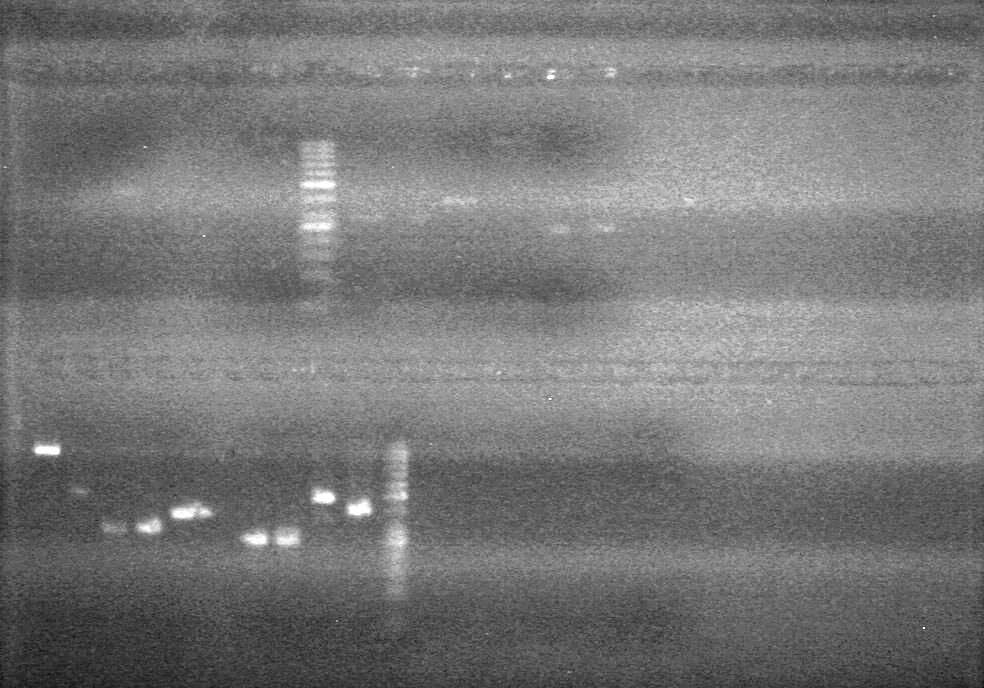
<b>Objective:</b> Ligate dT into pSB1C3.<br> | <b>Objective:</b> Ligate dT into pSB1C3.<br> | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 331: | Line 335: | ||
<tr><td>20<td>pLacI PCR product<td> good | <tr><td>20<td>pLacI PCR product<td> good | ||
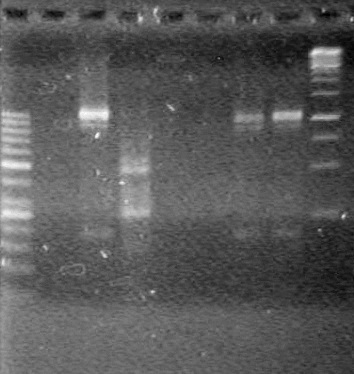
</table><br> | </table><br> | ||
| + | |||
| + | [[image:Lethbridge_100803AS.JPG|200px]] | ||
---- | ---- | ||
<b>Objective:</b> To ligate: pBAD-sRBS/mRBS, sRBS/mRBS-TetR, TetR-dT, dT-pTet, pLacI-sRBS/mRBS, Mms6-ptet28(a), dT-PSB1C3<br> | <b>Objective:</b> To ligate: pBAD-sRBS/mRBS, sRBS/mRBS-TetR, TetR-dT, dT-pTet, pLacI-sRBS/mRBS, Mms6-ptet28(a), dT-PSB1C3<br> | ||
| - | <b>Method:</b> [[Team:Lethbridge/Notebook/Protocols|Ligation of Plasmid DNA]] | + | <b>Method:</b> [[Team:Lethbridge/Notebook/Protocols|Ligation of Plasmid DNA]]<br> |
| - | + | 15µL pDNA in plasmid, and 15 µL of pDNA biobrick | |
==<font color="white">August 4, 2010== | ==<font color="white">August 4, 2010== | ||
| Line 392: | Line 398: | ||
*Ran at 100V for 70 minutes. | *Ran at 100V for 70 minutes. | ||
| - | <b>Results:</b> | + | <b>Results:</b>[[image:Lethbridge_100804 JV Ligation PCR AS.JPG|200px]] <br><br><br> |
| + | |||
| + | |||
| + | |||
<b>Objective:</b> Transform the successful ligations<br> | <b>Objective:</b> Transform the successful ligations<br> | ||
| Line 454: | Line 463: | ||
Gel ran for 60 minutes at 100 V. Small & faint band was slightly visible. | Gel ran for 60 minutes at 100 V. Small & faint band was slightly visible. | ||
| - | Gel extraction was carried out using QIAGEN method. Eluted to 12 (µL). | + | Gel extraction was carried out using QIAGEN method. Eluted to 12 (µL).<br> |
| + | [[image:Lethbridge_100809 JV Mms6 Gel Extract BW.jpg|200px]] | ||
==<font color="white">August 6, 2010 Evening== | ==<font color="white">August 6, 2010 Evening== | ||
| Line 522: | Line 532: | ||
<tr><td>7<td>lumazine(justin's)<td>5(µL) sample, 1(µL)dye | <tr><td>7<td>lumazine(justin's)<td>5(µL) sample, 1(µL)dye | ||
</table><br> | </table><br> | ||
| + | [[image:Lethbridge_100806ADSPCR.JPG|200px]] | ||
Repeat gel with template controls | Repeat gel with template controls | ||
| Line 537: | Line 548: | ||
<tr><td>1<td>1Kb ladder<td>0.5(µL) ladder, 2(µL) dye, 9.5(µL) H<sub>2</sub>0 | <tr><td>1<td>1Kb ladder<td>0.5(µL) ladder, 2(µL) dye, 9.5(µL) H<sub>2</sub>0 | ||
</table><br> | </table><br> | ||
| + | [[image:Lethbridge_100809AS-PCR.jpg|200px]] | ||
*ran at 100V for 75 minutes | *ran at 100V for 75 minutes | ||
| Line 570: | Line 582: | ||
</table><br> | </table><br> | ||
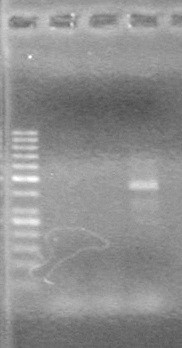
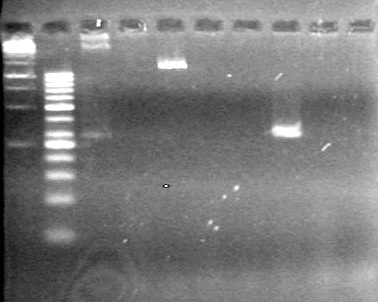
| - | Ran at 100 V for 45 minutes. mRBS-xylE did not amplify while lumazine did amplify. | + | Ran at 100 V for 45 minutes. mRBS-xylE did not amplify while lumazine did amplify. <br> |
| - | + | <b>Results:</b> Ligation of mRBS-xylE NOT confirmed<br> | |
| + | [[image:Lethbridge_100809 HB xylE Lum PCR BW.jpg|50px]] | ||
| + | |||
---- | ---- | ||
| Line 656: | Line 670: | ||
<tr><td>20<td>pLacI-sRBS 3 URD | <tr><td>20<td>pLacI-sRBS 3 URD | ||
</table><br> | </table><br> | ||
| + | [[image:Lethbridge_100809AVKG Top.jpg|200px]] | ||
<table border ="3"> | <table border ="3"> | ||
| Line 681: | Line 696: | ||
</table><br> | </table><br> | ||
| - | + | [[image:Lethbridge_100809AVKG Bottom.jpg|200px]] | |
---- | ---- | ||
| Line 690: | Line 705: | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| - | + | ||
| - | + | <u>Restrictions</u><br> | |
*Restrict Lumazine wit EcoRI and SpeI (Red Buffer)<br> | *Restrict Lumazine wit EcoRI and SpeI (Red Buffer)<br> | ||
*Restrict the dT with XbaI and EcoRI (Orange Buffer)<br> | *Restrict the dT with XbaI and EcoRI (Orange Buffer)<br> | ||
| Line 706: | Line 721: | ||
Incubated reactions for 60 minutes at 37<sup>o</sup>C<br> | Incubated reactions for 60 minutes at 37<sup>o</sup>C<br> | ||
| - | + | <u>Ligation</u><br> | |
Reaction set up as follows: | Reaction set up as follows: | ||
*T4 DNA ligase - 0.25µL<br> | *T4 DNA ligase - 0.25µL<br> | ||
| Line 771: | Line 786: | ||
</table><br> | </table><br> | ||
| - | + | [[image:Lethbridge_100810ReRunRestrictionDigers.JPG|200px]] | |
---- | ---- | ||
| Line 814: | Line 829: | ||
</table><br> | </table><br> | ||
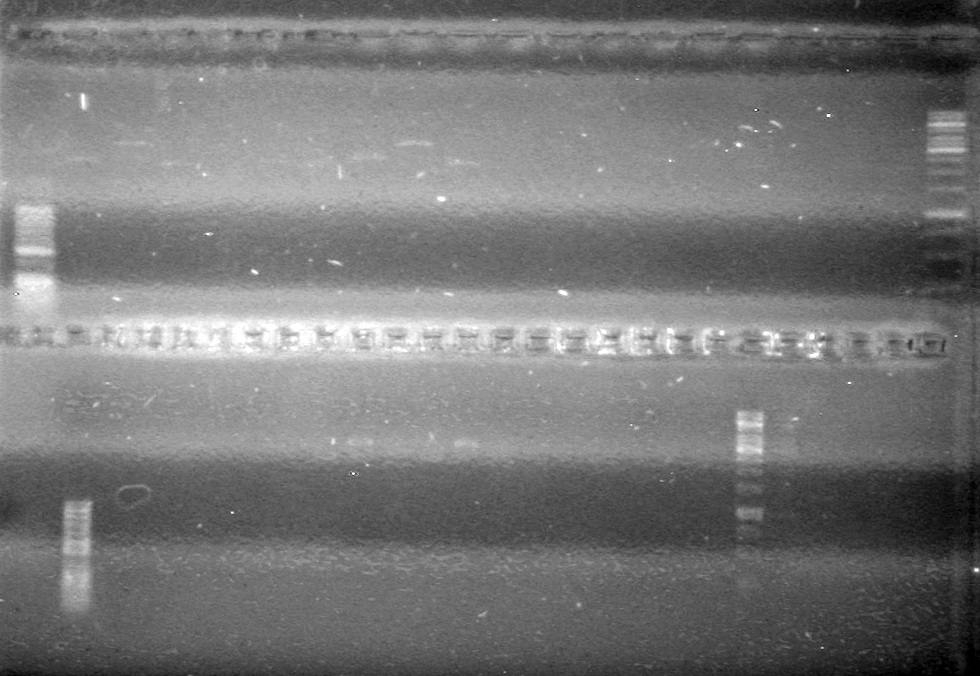
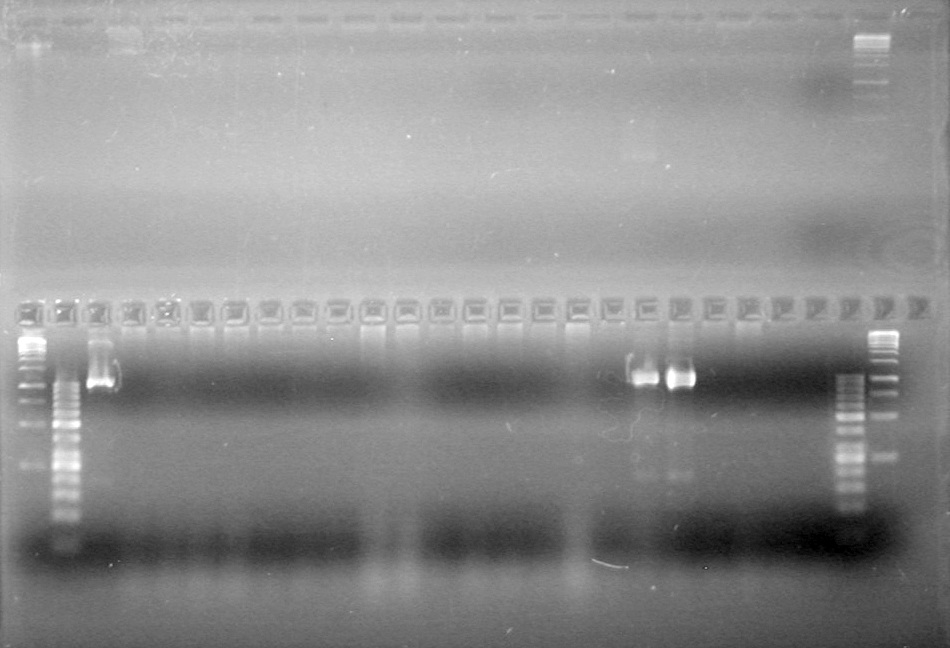
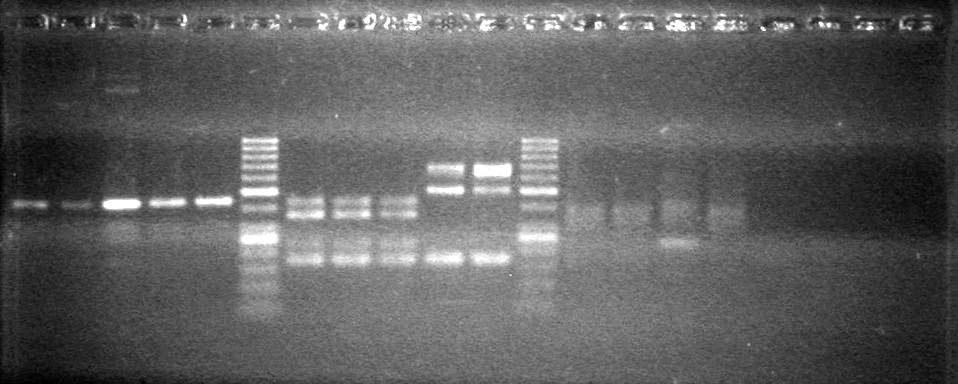
| - | Controls: sRBS, pTET & mRBS - will PCR these to compare size using same conditions as above. | + | Controls: sRBS, pTET & mRBS - will PCR these to compare size using same conditions as above. <br> |
| - | + | [[image:Lethbridge_100810MassiveColonyPCR JV AS.JPG|200px]]<br> | |
| - | + | [[image:Lethbridge_100810MassiveColonyPCR Large Gel JV AS.JPG|200px]] | |
==<font color="white">Aug 10, 2010 Evening== | ==<font color="white">Aug 10, 2010 Evening== | ||
| Line 847: | Line 862: | ||
</table><br> | </table><br> | ||
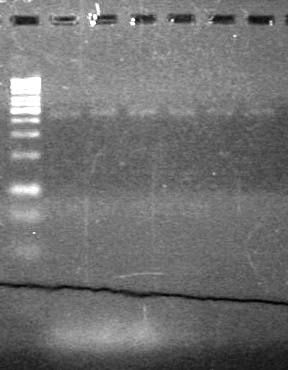
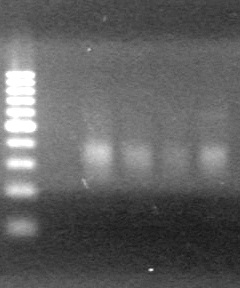
| - | Ran samples on 1.5% agarose gel (1X TAE) for 60 minutes at 100V. | + | Ran samples on 1.5% agarose gel (1X TAE) for 60 minutes at 100V. <br> |
| - | + | <b>Results:</b><br> | |
| + | [[image:Lethbridge_100810xylE PCR AS.JPG|100px]] | ||
==<font color="white">Aug 11, 2010== | ==<font color="white">Aug 11, 2010== | ||
| Line 870: | Line 886: | ||
<tr><td>dNTPs<td>2<td>15 | <tr><td>dNTPs<td>2<td>15 | ||
<tr><td>Forward Primer (VF2)<td>2<td>15 | <tr><td>Forward Primer (VF2)<td>2<td>15 | ||
| - | <tr><td>Reverse | + | <tr><td>Reverse Primer (VR)<td>2<td>15 |
<tr><td>Template DNA<td>2<td> | <tr><td>Template DNA<td>2<td> | ||
<tr><td>Pfu polymerase<td>0.2<td>1.5 | <tr><td>Pfu polymerase<td>0.2<td>1.5 | ||
| Line 907: | Line 923: | ||
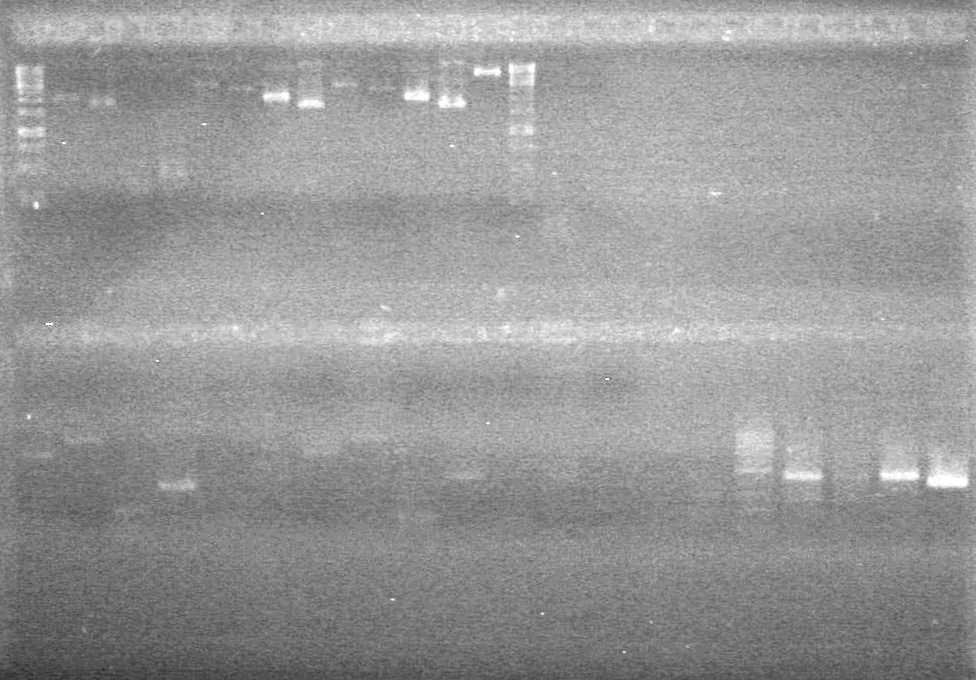
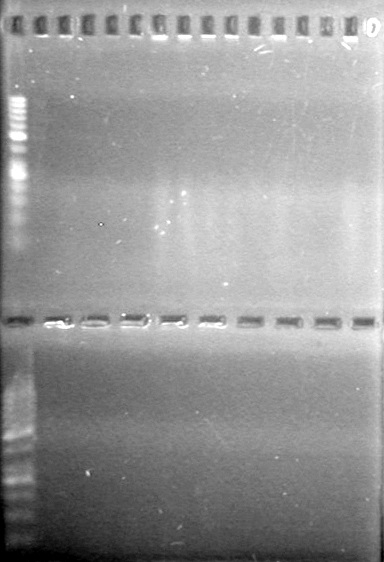
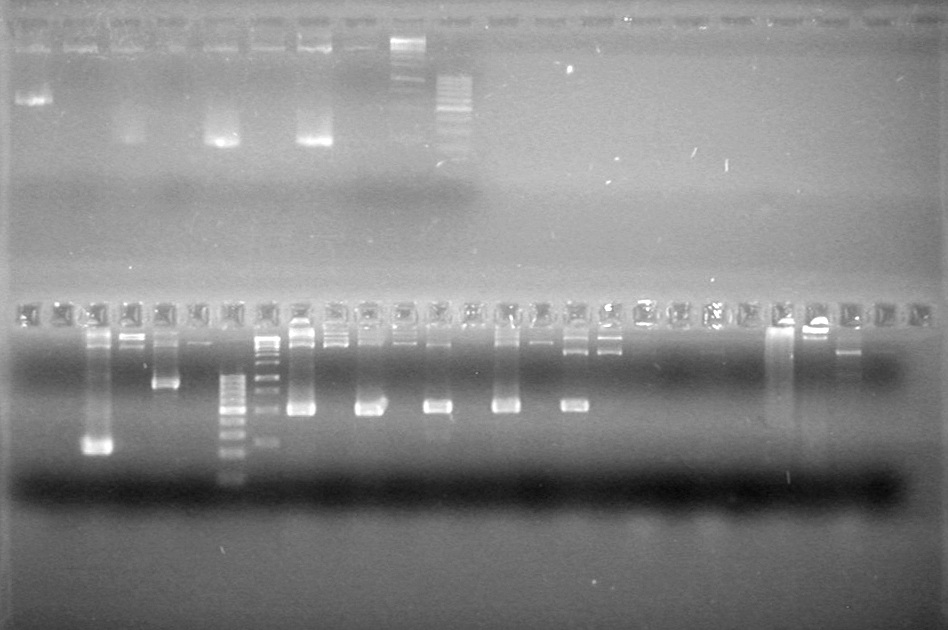
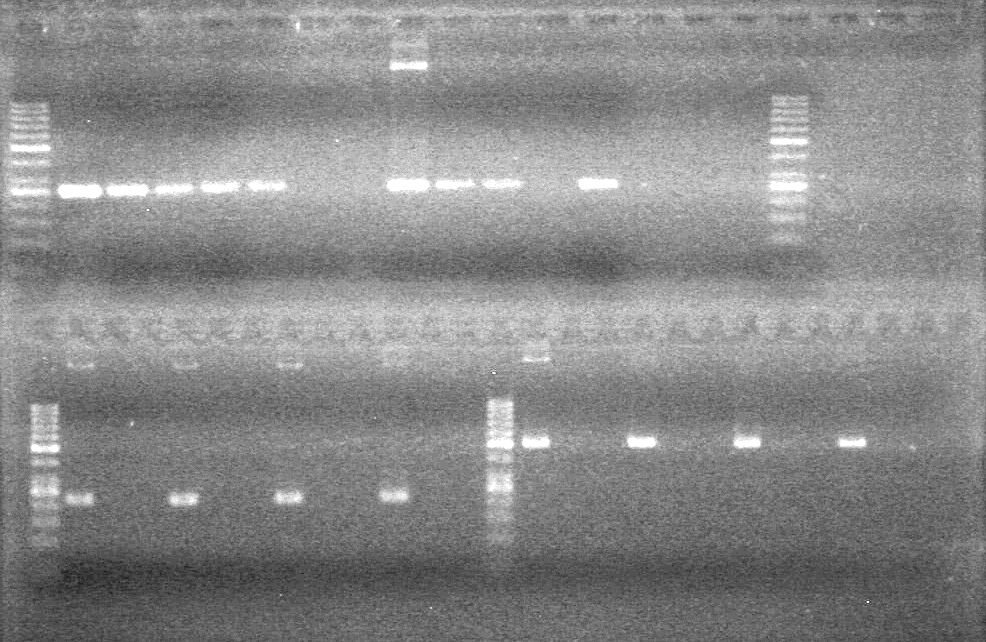
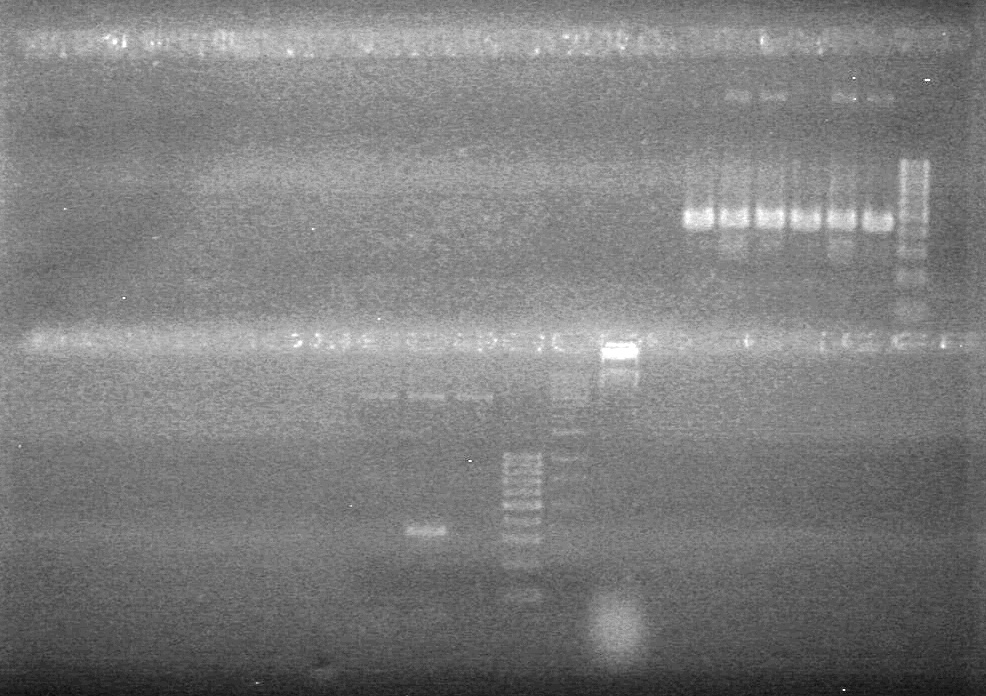
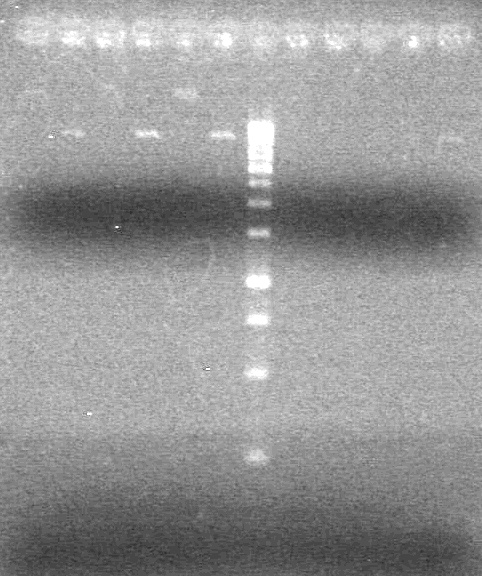
<b>Results:</b> Lanes 2, 3, 4 and 8 showed PCR amplification. Colonies chosen don't show the correct insert size. <br> | <b>Results:</b> Lanes 2, 3, 4 and 8 showed PCR amplification. Colonies chosen don't show the correct insert size. <br> | ||
| + | [[image:Lethbridge_100812 JV AS Colony PCR Pfu.JPG|200px]] | ||
| + | |||
| + | ---- | ||
| + | (In Lab: JV)<br> | ||
| + | <b>Objective:</b> Screen for colonies with the correct insert from Aug. 4, 2010 transformations. <br> | ||
| + | |||
| + | <b>Method:</b> Colony PCR. Changes - Used pipette tip instead of toothpick. Put colony in 20(µL) autoclaved Milli-Q water. <br> | ||
| + | <u>PCR:</u> Thermocycler set to iGEM PFUTEST<br> | ||
| + | |||
| + | 1. pLacI-sRBS: A (11 - 17) | ||
| + | 2. pBAD-sRBS: B (11 - 17) | ||
| + | 3. dT-pTet: C (11 - 17) | ||
| + | 4. pLacI-mRBS: D (11-17) | ||
| + | 5. pBAD-mRBS: E (11-17) | ||
| + | 6. mRBS-TetR: F (11-17) | ||
| + | |||
| + | Controls - mRBS, pTet, sRBS | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x47)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>9.8<td>460.6 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>94 | ||
| + | <tr><td>dNTPs<td>2<td>94 | ||
| + | <tr><td>Forward Primer (VF2)<td>2<td>94 | ||
| + | <tr><td>Reverse Primer (VR)<td>2<td>94 | ||
| + | <tr><td>Template DNA (Cell Lysate)<td>2<td>94 | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>9.4 | ||
| + | </table><br> | ||
| + | |||
| + | Added 18µL Master Mix to each reaction tube.<br> | ||
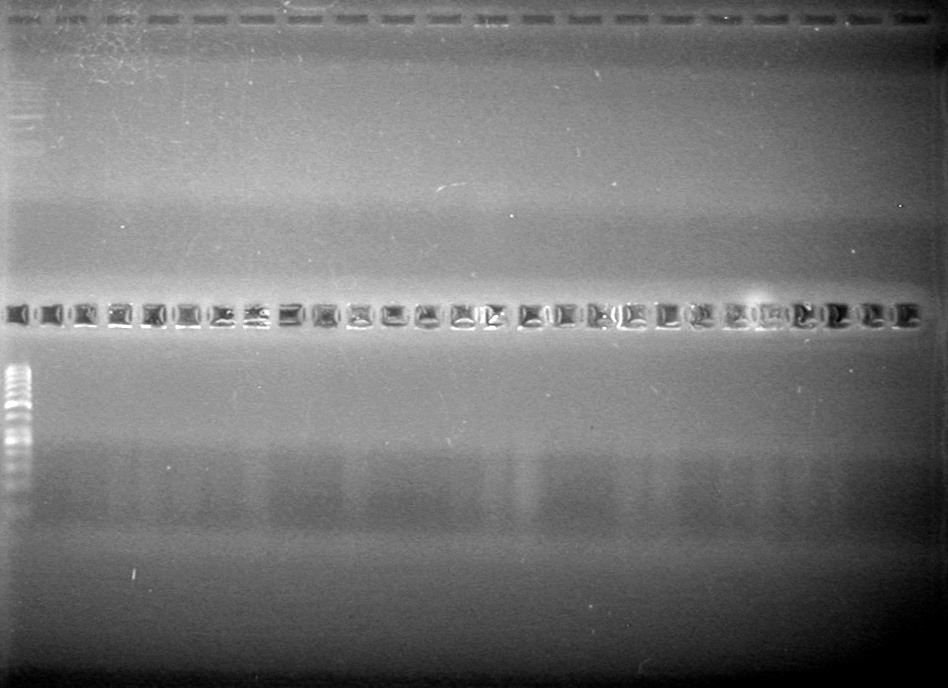
| + | Analyzed PCR products on 2.5% TAE gel.<br> | ||
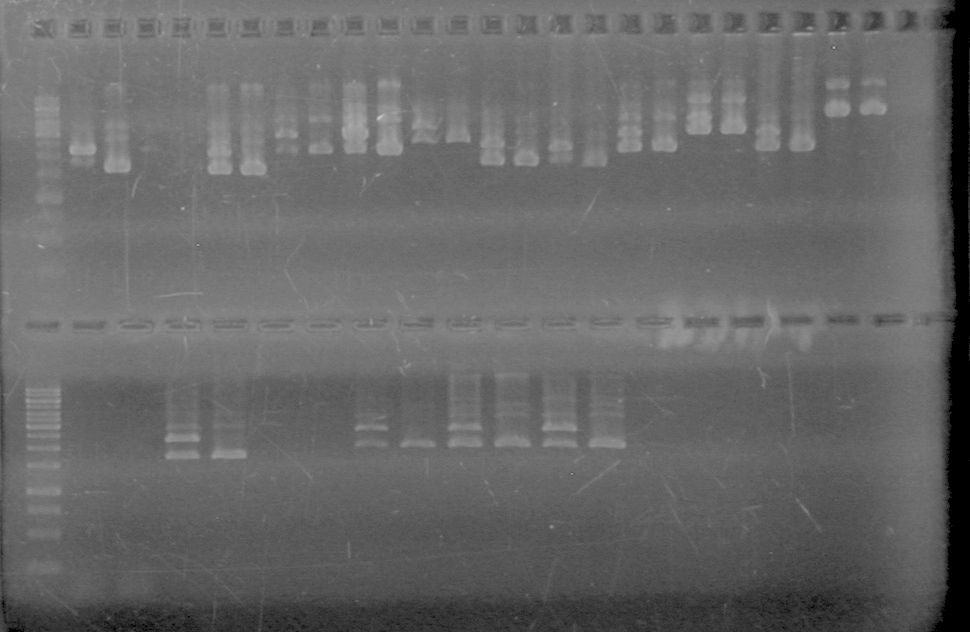
| + | <b>Results:</b><br> | ||
| + | [[image:Lethbridge_100812ASJVMassColonyPCRLargeGel (1).JPG|200px]]<br><br> | ||
| + | [[image:Lethbridge_100812ASJVMassColonyPCRSmallGel.JPG|200px]] | ||
| + | |||
| + | ---- | ||
| + | (In Lab: ADS, KG)<br> | ||
| + | <b>Objective:</b> Perform PCR on lumazine, mms6, xylE plasmids with prefix and suffix primers (these will tell us exact size without subtracting VF2/VR regions). If right will ligate into pET28a plasmids. <br> | ||
| + | |||
| + | <b>Method:</b> Plasmids used included: 6 mms6 maxipreps. 4 lumazine maxipreps. 5 xylE maxipreps. 1 mRBS maxiprep<br> | ||
| + | <u>PCR:</u> Thermocycler set to iGEM Program #11 PFU - P/S<br> | ||
| + | |||
| + | PCR- Conditions: | ||
| + | 1. Initial Denaturation 95<sup>o</sup>C for 3 min | ||
| + | 2. Denaturation 95<sup>o</sup>C for 30 sec | ||
| + | 3. Anneal (54<sup>o</sup>C) for 30 sec | ||
| + | 4. Extend 72<sup>o</sup>C for 3 min | ||
| + | 5. Final Extend 72<sup>o</sup>C for 15 min | ||
| + | 6. Held 4<sup>o</sup>C infinitely | ||
| + | (25 cycles) | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x16.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>9.8<td>161.7 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>33 | ||
| + | <tr><td>dNTPs<td>2<td>33 | ||
| + | <tr><td>Forward Primer (Prefix)<td>2<td>33 | ||
| + | <tr><td>Reverse Primer (Suffix Antisense)<td>2<td>33 | ||
| + | <tr><td>Template DNA (Cell Lysate)<td>2<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>3.3 | ||
| + | </table><br> | ||
| + | |||
| + | Added 18µL Master Mix to each reaction tube.<br> | ||
| + | Analyzed PCR products on 2.5% TAE gel run at 100 V for 35 minutes.<br> | ||
| + | [[image:Lethbridge_100813ADS Awesome PCR.JPG|200px]] | ||
==<font color="white">Aug 13, 2010== | ==<font color="white">Aug 13, 2010== | ||
| Line 921: | Line 1,002: | ||
<tr><td>dNTPs<td>1<td>12.5 | <tr><td>dNTPs<td>1<td>12.5 | ||
<tr><td>Forward Primer (VF2)<td>0.5<td>6.25 | <tr><td>Forward Primer (VF2)<td>0.5<td>6.25 | ||
| - | <tr><td>Reverse | + | <tr><td>Reverse Primer (VR)<td>0.5<td>6.25 |
<tr><td>Template DNA<td>1<td> | <tr><td>Template DNA<td>1<td> | ||
<tr><td>Pfu polymerase<td>0.15<td>1.888 | <tr><td>Pfu polymerase<td>0.15<td>1.888 | ||
</table><br> | </table><br> | ||
| - | Added 49µL Master Mix to each reaction tube. | + | Added 49µL Master Mix to each reaction tube.<br> |
| + | |||
| + | |||
| + | ---- | ||
| + | (In Lab: AS)<br> | ||
| + | |||
| + | <b>Objective:</b> PCR amplify pSB1A3, pSB1T3 and pSB1C3 for use in future 3 part assembly and subsequent growth for glycerol stock. <br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x13.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>14.9<td>52.15 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>7 | ||
| + | <tr><td>dNTPs<td>1<td>3.5 | ||
| + | <tr><td>Forward Primer (SB-prep-2)<td>0.7<td>2.45 | ||
| + | <tr><td>Reverse Primer (SB-prep-3p)<td>0.7<td>2.45 | ||
| + | <tr><td>Template DNA<td>0.5<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>0.7 | ||
| + | </table><br> | ||
| + | |||
| + | PCR- Conditions: | ||
| + | 1. 94<sup>o</sup>C for 30 sec | ||
| + | 2. 94<sup>o</sup>C for 30 sec | ||
| + | 3. 55<sup>o</sup>C for 30 sec | ||
| + | 4. 68<sup>o</sup>C for 3 min | ||
| + | 5. 68<sup>o</sup>C for 10 min | ||
| + | 6. 4<sup>o</sup>C infinitely | ||
| + | (36 cycles) | ||
| + | |||
| + | Added 19.5µL Master Mix to each reaction tube.<br> | ||
| + | Analyzed products on 1% TAE agarose gel which ran for 60 minutes at 100 V.<br> | ||
| + | |||
| + | <b>Results:</b><br> | ||
| + | [[image:Lethbridge_100814PlasmidBackbones.JPG|100px]] | ||
==<font color="white">Aug 14, 2010== | ==<font color="white">Aug 14, 2010== | ||
| Line 955: | Line 1,068: | ||
<tr><td>19<td>MT | <tr><td>19<td>MT | ||
<tr><td>20<td>MT | <tr><td>20<td>MT | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</table> | </table> | ||
| Line 995: | Line 1,101: | ||
</table> | </table> | ||
| - | + | [[image:Lethbridge_100815MiniprepPCR.JPG|200px]] | |
| + | |||
| + | ---- | ||
| + | (In Lab: AS)<br> | ||
| + | |||
| + | <b>Objective:</b> Insert mms6 and lumazine into pET28a using NotI restriction site. <br> | ||
| + | |||
| + | <b>Method:</b> 1. Reserve 1(µL) of dirty PCR product for analysis. 2. Clean up PCR products using Qiagen prep. 3. Restrict 4 mms6 maxipreps and 4 lumazine maxipreps.<br> | ||
| + | |||
| + | <u>Restriction Reactions:</u> | ||
| + | |||
| + | For lumazine and mms6 - | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>1X(µL)</b><td><b>Master Mix(x8.5)(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>7.8<td>66.30 | ||
| + | <tr><td>Orange Buffer (10x)</td><td>2<td>17 | ||
| + | <tr><td>pDNA</td><td>10<td> | ||
| + | <tr><td>NotI</td><td>0.2<td>1.7 | ||
| + | </table> | ||
| + | |||
| + | Added 10 (µL) to each tube. | ||
| + | |||
| + | For pET28a - | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>Reaction Mix(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>4.8 | ||
| + | <tr><td>Orange Buffer (10x)</td><td>5 | ||
| + | <tr><td>pDNA</td><td>40 | ||
| + | <tr><td>NotI</td><td>0.2 | ||
| + | </table> | ||
| + | |||
| + | Incubated at 37<sup>o</sup>C. | ||
| + | Analyzed results on 2.5% TAE agarose gel which ran at 100 V for 50 minutes.<br> | ||
| + | [[image:Lethbridge_100814ColonyPCRRetryandStartofassembly.JPG|200px]] | ||
| + | |||
| + | <b>Results:</b> Lost all the DNA in the column clean-up step and will have to re-do.<br> | ||
==<font color="white">Aug 14, 2010 Evening== | ==<font color="white">Aug 14, 2010 Evening== | ||
| Line 1,003: | Line 1,144: | ||
<b>Objective:</b> Assemble mms6-dT and lumazine-dT using three antibiotic assembly. <br> | <b>Objective:</b> Assemble mms6-dT and lumazine-dT using three antibiotic assembly. <br> | ||
| - | <b>Method:</b> | + | <b>Method:</b> |
| + | #PCR amplify BioBricks (Prefix/Suffix) | ||
| + | #Restrict BioBricks | ||
| + | #Ligate BioBricks into psB1C3 | ||
| + | #Confirm ligation by PCR analysis (VF2/VR) | ||
| + | #Transform ligation mixes | ||
| + | #Screen colonies with Colony PCR | ||
<u>PCR:</u> Thermocycler set to iGEM program 11<br> | <u>PCR:</u> Thermocycler set to iGEM program 11<br> | ||
| Line 1,043: | Line 1,190: | ||
</table> | </table> | ||
| - | For | + | For pSB1C3 - |
<table><table border ="3"> | <table><table border ="3"> | ||
<tr><td><b>Ingredient</b><td><b>Reaction Mix(µL)</b> | <tr><td><b>Ingredient</b><td><b>Reaction Mix(µL)</b> | ||
| Line 1,131: | Line 1,278: | ||
</table><br> | </table><br> | ||
| - | Added 15 (µL) MM to each tube. | + | Added 15 (µL) MM to each tube.<br> |
| + | <b>Results:</b><br> | ||
| + | |||
| + | ==<font color="white">Aug 15, 2010== | ||
| + | (In Lab: ADS)<br> | ||
| + | |||
| + | <b>Objective:</b> Insert mms6 and lumazine into pET28a using NotI restriction site. <br> | ||
| + | |||
| + | <b>Method:</b> 1. Reserve 1(µL) of dirty PCR product for analysis. 2. Clean up PCR products using Qiagen prep. 3. Restrict 3 mms6 maxipreps and 2 lumazine maxipreps.<br> | ||
| + | |||
| + | <u>Restriction Reactions:</u> | ||
| + | |||
| + | For lumazine and mms6 - | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>1X(µL)</b><td><b>Master Mix(x10.5)(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>11.8<td>123.9 | ||
| + | <tr><td>Orange Buffer (10x)</td><td>2<td>21 | ||
| + | <tr><td>pDNA</td><td>6<td> | ||
| + | <tr><td>NotI</td><td>0.2<td>2.1 | ||
| + | </table> | ||
| + | |||
| + | Added 14 (µL) to each tube. | ||
| + | |||
| + | For pET28a - | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>Reaction Mix(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>4.8 | ||
| + | <tr><td>Orange Buffer (10x)</td><td>5 | ||
| + | <tr><td>pDNA</td><td>40 | ||
| + | <tr><td>NotI</td><td>0.2 | ||
| + | </table> | ||
| + | |||
| + | Incubated at 37<sup>o</sup>C for 1.5 hours. Heat killed enzymes at 80<sup>o</sup>C for 20 minutes. | ||
| + | |||
| + | <u>Ligation Reactions:</u> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>1X(µL)</b><td><b>Master Mix(x5.5)(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>13.9<td>75.9 | ||
| + | <tr><td>T4 Ligase Buffer (10x)</td><td>2<td>11 | ||
| + | <tr><td>Plasmid (pET28a)</td><td>2<td>11 | ||
| + | <tr><td>Cut out part (Lumazine/mms6)</td><td>2<td> | ||
| + | <tr><td>T4 DNA Ligase</td><td>0.2<td>1.1 | ||
| + | </table> | ||
| + | |||
| + | Added 18(µL) to each tube. Incubated 1 hour and overnight at room temperature. | ||
==<font color="white">Aug 15, 2010 Evening== | ==<font color="white">Aug 15, 2010 Evening== | ||
| Line 1,192: | Line 1,384: | ||
Added 45 (µL) MM to each tube. | Added 45 (µL) MM to each tube. | ||
| - | Analyzed PCR products of BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. | + | Analyzed PCR products of BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. <br> |
| + | <b>Results:</b><br> | ||
| + | [[image:Lethbridge_100815PCRAssembly.JPG|200px]] | ||
| - | + | ||
| + | ---- | ||
| + | (In Lab: ADS)<br> | ||
| + | |||
| + | <b>Objective:</b> Produce large quantities of pSB1A3, pSB1C3 and pSB1T3.<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x3.2)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>75<td>240 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>10<td>32 | ||
| + | <tr><td>dNTPs<td>5<td>16 | ||
| + | <tr><td>Primer (SB-prep-2Ea)<td>3.5<td>11.2 | ||
| + | <tr><td>Primer (SB-prep-3P)<td>3.5<td>11.2 | ||
| + | <tr><td>Template DNA<td>2<td> | ||
| + | <tr><td>Pfu polymerase<td>1<td>3.2 | ||
| + | </table><br> | ||
| + | |||
| + | Added 98(µL) to each tube. Used PLASRB PCR Protocol from Aug. 13, 2010. | ||
==<font color="white">Aug 16, 2010 == | ==<font color="white">Aug 16, 2010 == | ||
| Line 1,241: | Line 1,453: | ||
</table><br> | </table><br> | ||
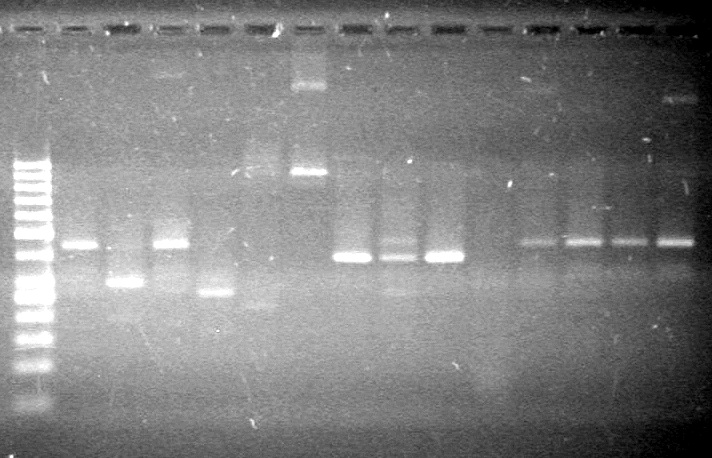
| - | Analyzed PCR products of overnight BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. | + | Analyzed PCR products of overnight BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. <br> |
| - | + | <b>Results:</b><br> | |
| - | + | [[image:Lethbridge_100816PostLigationPCRScreen.JPG|200px]] | |
==<font color="white">Aug 16, 2010 Evening == | ==<font color="white">Aug 16, 2010 Evening == | ||
| Line 1,249: | Line 1,461: | ||
(In Lab: KG, AS)<br> | (In Lab: KG, AS)<br> | ||
| - | <b>Objective:</b> Restriction of PCR Products (mms6-dT, lumazine-dT). Restriction is necessary for ligation into plasmid backbone | + | <b>Objective:</b> Restriction of PCR Products (mms6-dT, lumazine-dT). Restriction is necessary for ligation into plasmid backbone pSB1C3 <br> |
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 1,275: | Line 1,487: | ||
<tr><td>T4 DNA Ligase</td><td>0.2<td>1.1 | <tr><td>T4 DNA Ligase</td><td>0.2<td>1.1 | ||
</table> | </table> | ||
| + | |||
| + | ---- | ||
| + | (In Lab: KG)<br> | ||
| + | |||
| + | <b>Objective:</b> Transformations of insertions of mms6 or lumazine into pET28a. <br> | ||
| + | |||
| + | <b>Method:</b> used [[Team:Lethbridge/Notebook/Protocols|Competent Cell Transformation]] protocol | ||
| + | * changes: | ||
| + | **used 50µL aliquottes of DH5&alpha | ||
| + | **did not pipette up and down once, the cells were just swirled 3 times | ||
| + | **added 400µL SOC media, shoock at 37<sup>0</sup>C for 90 min | ||
| + | **platted 250µL and 150µL<br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <td><b>results</b> | ||
| + | <tr><td>contents<td><b>&250µL</b><td><b>150µL</b> | ||
| + | <tr><td>+ control(pUC19)<td>good<td>good | ||
| + | <tr><td>mms6<td>good<td>good | ||
| + | <tr><td>mms6-2<td>good<td>good | ||
| + | <tr><td>mms6<td>good<td>x | ||
| + | <tr><td>Lumazine<td>good<td>x | ||
| + | <tr><td>Lumazine<td>good<td>x | ||
| + | </table><br> | ||
==<font color="white">Aug 17, 2010 == | ==<font color="white">Aug 17, 2010 == | ||
| Line 1,296: | Line 1,531: | ||
</table><br> | </table><br> | ||
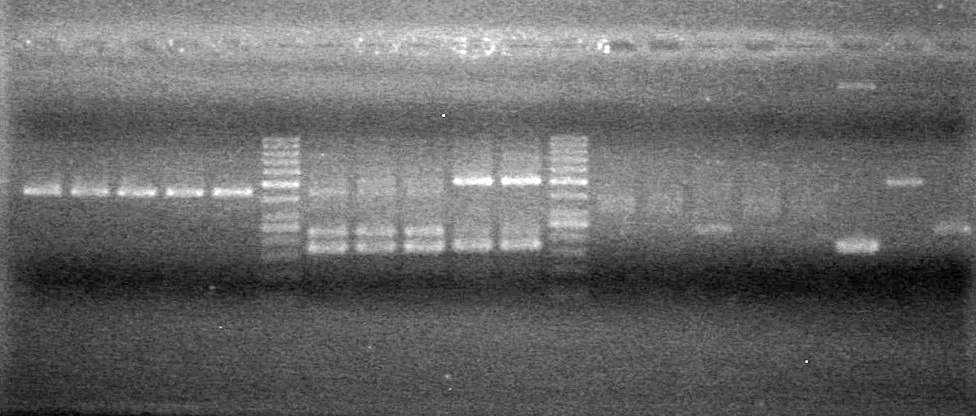
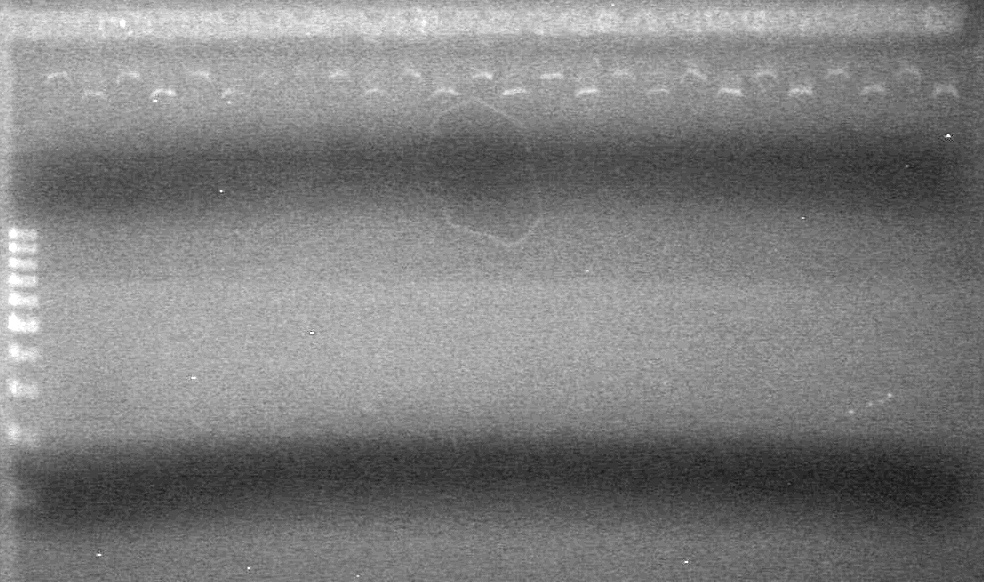
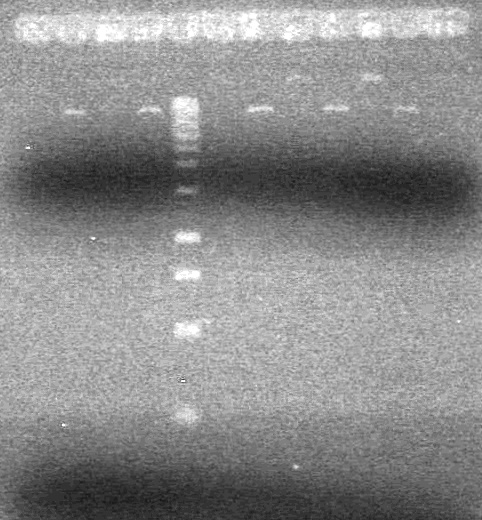
| - | Ran a 2% Agarose gel in 1X TAE buffer for 65 minutes at 100V. | + | Ran a 2% Agarose gel in 1X TAE buffer for 65 minutes at 100V.<br> |
| - | + | [[image:Lethbridge_100817PostLigationPCRTest.JPG|100px]] | |
| - | + | ||
==<font color="white">Aug 17, 2010 Evening == | ==<font color="white">Aug 17, 2010 Evening == | ||
| Line 1,363: | Line 1,597: | ||
Added 45(µL) MM to each rxn tube. | Added 45(µL) MM to each rxn tube. | ||
| + | ---- | ||
| + | <b>Objective:</b> Analyze tendency of PstI to be heat killed. <br> | ||
| + | <b>Hypothesis:</b> Ligation will be counteracted by PstI digestion even following 20 minute heat kill at 80<sup>o</sup>C .<br> | ||
| + | <b>Method:</b> Restrict plasmid DNA with PstI (and EcoRI control). Reactions stopped by heat killing. Plasmids re-ligated with T4 DNA Ligase. Ligation confirmed with PCR. If PCR product is present then enzyme heat killed but if there is no PCR Product then enzyme was not heat killed. <br> | ||
| + | |||
| + | <u>Restriction Reactions:</u> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>1X(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>12.8 | ||
| + | <tr><td>Orange Buffer (10x)</td><td>2 | ||
| + | <tr><td>pDNA (dT)</td><td>5 | ||
| + | <tr><td>PstI</td><td>0.2 | ||
| + | <tr><td>EcoRI (control only)</td><td>0.2 | ||
| + | </table> | ||
| + | |||
| + | Incubated at 37<sup>o</sup>C for 1.5 hours. Heat killed enzymes for 20 minutes at 80<sup>o</sup>C. | ||
| + | |||
| + | <u>Ligation Reactions:</u> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>1X(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>15.8 | ||
| + | <tr><td>T4 Ligase Buffer (10x)</td><td>2 | ||
| + | <tr><td>Restriction Mix</td><td>2 | ||
| + | <tr><td>T4 DNA Ligase</td><td>0.2 | ||
| + | </table> | ||
| + | |||
| + | Incubated at room temperature overnight. | ||
| + | |||
| + | PCR to confirm Ligation | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x2.2)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>33.8<td>74.36 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>5<td>11 | ||
| + | <tr><td>dNTPs<td>2<td>4.4 | ||
| + | <tr><td>Forward VF2 Primer<td>2<td>4.4 | ||
| + | <tr><td>Reverse VR Primer<td>2<td>4.4 | ||
| + | <tr><td>Template DNA<td>5<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>0.44 | ||
| + | </table><br> | ||
| + | |||
| + | Added 45(µL) to each reaction tube. | ||
| + | |||
| + | ==<font color="white">Aug 18, 2010 == | ||
| + | |||
| + | (In Lab: JV)<br> | ||
| + | <b>Objective:</b> Confirmation of ADS' analysis of PstI's tendency to be heat killed. <br> | ||
| + | <b>Method:</b> Will PCR ligation reactions from different assembly methods and EcoRI and PstI controls. <br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>33.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>5 | ||
| + | <tr><td>dNTPs<td>2 | ||
| + | <tr><td>Forward VF2 Primer<td>2 | ||
| + | <tr><td>Reverse VR Primer<td>2 | ||
| + | <tr><td>Template DNA<td>5 | ||
| + | <tr><td>Pfu polymerase<td>0.2 | ||
| + | </table><br> | ||
| + | Ran on cycle 4 of thermocycler. | ||
| + | |||
| + | Viewed PCRs on 2% TAE agarose gel that ran at 110 V for 30 minutes.<br> | ||
| + | [[image:Lethbridge_100818AssemblyandHeatKillPCR.JPG|200px]] | ||
| + | ---- | ||
| + | (In Lab: HB)<br> | ||
| + | <b>Objective:</b> Analyze tendency of PstI to be heat killed. <br> | ||
| + | <b>Method:</b> Restrict plasmid DNA with PstI (and EcoRI control). Reactions stopped by heat killing. Plasmids re-ligated with T4 DNA Ligase. Ligation confirmed with PCR. If PCR product is present then enzyme heat killed but if there is no PCR Product then enzyme was not heat killed. <br> | ||
| + | |||
| + | <u>Restriction Reactions:</u> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>1X(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>12.8 | ||
| + | <tr><td>Orange Buffer (10x)</td><td>2 | ||
| + | <tr><td>pDNA (dT)</td><td>5 | ||
| + | <tr><td>PstI</td><td>0.2 | ||
| + | <tr><td>EcoRI (control only)</td><td>0.2 | ||
| + | </table> | ||
| + | |||
| + | Incubated at 37<sup>o</sup>C for 1.5 hours. Heat killed enzymes for 20 minutes at 80<sup>o</sup>C. | ||
| + | |||
| + | <u>Ligation Reactions:</u> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Ingredient</b><td><b>1X(µL)</b> | ||
| + | <tr><td>MilliQ H<sub>2</sub>0 Water</td><td>15.8 | ||
| + | <tr><td>T4 Ligase Buffer (10x)</td><td>2 | ||
| + | <tr><td>Restriction Mix</td><td>2 | ||
| + | <tr><td>T4 DNA Ligase</td><td>0.2 | ||
| + | </table> | ||
| + | |||
| + | Incubated at room temperature overnight. | ||
| + | |||
| + | PCR to Confirm Ligation | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x6.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>33.8<td>219.7 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>5<td>32.5 | ||
| + | <tr><td>dNTPs<td>2<td>13 | ||
| + | <tr><td>Forward VF2 Primer<td>2<td>13 | ||
| + | <tr><td>Reverse VR Primer<td>2<td>13 | ||
| + | <tr><td>Template DNA<td>5<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>1.3 | ||
| + | </table><br> | ||
| + | |||
| + | Added 45(µL) to each reaction tube. | ||
| + | Used thermocycler iGEM Program 4. | ||
| + | |||
| + | 3 different treatments: 1. Restricted, heat killed and ligated. 2. Restricted and heat killed. 3. Restricted and not heat killed. | ||
==<font color="white">Aug 19, 2010 == | ==<font color="white">Aug 19, 2010 == | ||
| Line 1,411: | Line 1,751: | ||
Samples were run on a 2% agarose gel in 1X TAE Buffer. | Samples were run on a 2% agarose gel in 1X TAE Buffer. | ||
| - | + | ---- | |
| + | (In Lab: HB)<br> | ||
| + | |||
| + | <b>Objective:</b> Run a PCR testing VF2/Suffix and Prefix/VR. A colony PCR of lumazine and mms6 in pET28a. A PCR of 15 ligation tests. <br> | ||
| + | (In Lab: JV)<br> | ||
| + | <b>Objective:</b> Screen for colonies with the correct insert from Aug. 4, 2010 transformations. <br> | ||
| + | |||
| + | <b>Method:</b> Colony PCR. Changes - Used pipette tip instead of toothpick. Put colony in 20(µL) autoclaved Milli-Q water. <br> | ||
| + | |||
| + | <u>PCR:</u> Thermocycler set to iGEM Program 4<br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>9.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2 | ||
| + | <tr><td>dNTPs<td>2 | ||
| + | <tr><td>Forward Primer (VF2)<td>2 | ||
| + | <tr><td>Reverse Primer (VR)<td>2 | ||
| + | <tr><td>Template DNA (Cell Lysate)<td>2 | ||
| + | <tr><td>Pfu polymerase<td>0.2 | ||
| + | </table><br> | ||
| + | |||
| + | Added 18µL Master Mix to each reaction tube. | ||
| + | |||
| + | <b>Method:</b> Control test of primer combinations. <br> | ||
| + | |||
| + | Combination 1 - VF2/Suffix | ||
| + | Combination 2 - Prefix/VR | ||
| + | Combination 3 - Prefix/Suffix | ||
| + | dT maxiprep used as known template DNA source. | ||
| + | |||
| + | <u>PCR Combination 1:</u><br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>9.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2 | ||
| + | <tr><td>dNTPs<td>2 | ||
| + | <tr><td>Forward Primer (Prefix)<td>2 | ||
| + | <tr><td>Reverse Primer (Suffix)<td>2 | ||
| + | <tr><td>Template DNA (dT)<td>2 | ||
| + | <tr><td>Pfu polymerase<td>0.2 | ||
| + | </table><br> | ||
| + | |||
| + | <u>PCR Combination 2:</u><br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>9.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2 | ||
| + | <tr><td>dNTPs<td>2 | ||
| + | <tr><td>Forward Primer (Prefix)<td>2 | ||
| + | <tr><td>Reverse Primer (VR)<td>2 | ||
| + | <tr><td>Template DNA (dT)<td>2 | ||
| + | <tr><td>Pfu polymerase<td>0.2 | ||
| + | </table><br> | ||
| + | |||
| + | <u>PCR Combination 3:</u><br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>9.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2 | ||
| + | <tr><td>dNTPs<td>2 | ||
| + | <tr><td>Forward Primer (VF2)<td>2 | ||
| + | <tr><td>Reverse Primer (suffix)<td>2 | ||
| + | <tr><td>Template DNA (dT)<td>2 | ||
| + | <tr><td>Pfu polymerase<td>0.2 | ||
| + | </table><br> | ||
| + | |||
| + | <b>Method:</b> PCR of 15 ligation tests. <br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x19.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>9.8<td>191.1 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>39 | ||
| + | <tr><td>dNTPs<td>2<td>39 | ||
| + | <tr><td>Forward VF2 Primer<td>2<td>39 | ||
| + | <tr><td>Reverse VR Primer<td>2<td>39 | ||
| + | <tr><td>Template DNA<td>2<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>3.9 | ||
| + | </table><br> | ||
| + | |||
| + | Added 18(µL) to each tube. | ||
| + | ---- | ||
| + | (In Lab: JV)<br> | ||
| + | Analyzed HB's PCR products on 2% TAE gel which ran for 60 minutes at 100 V.<br> | ||
| + | [[image:Lethbridge_100819PCRMania.JPG|200px]] | ||
==<font color="white">Aug 20, 2010== | ==<font color="white">Aug 20, 2010== | ||
| Line 1,423: | Line 1,846: | ||
<b>Results:</b> DNA concentration was good, however there was no evidence of an insert into pET-28(A). <br> | <b>Results:</b> DNA concentration was good, however there was no evidence of an insert into pET-28(A). <br> | ||
| - | + | [[image:Lethbridge_100819pET28aTransformScreenGel1.JPG|200px]]<br><br> | |
| + | [[image:Lethbridge_100819pET28aTransformScreenGel2.JPG|200px]]<br><br> | ||
| + | [[image:Lethbridge_100819pET28aTransformScreenGel3.JPG|200px]]<br><br> | ||
| + | [[image:Lethbridge_100819pET28aTransformScreenGel4.JPG|200px]]<br><br> | ||
==<font color="white">Aug 23, 2010== | ==<font color="white">Aug 23, 2010== | ||
| Line 1,459: | Line 1,885: | ||
</table> | </table> | ||
| - | 15(µL) added to each rxn tube. | + | 15(µL) added to each rxn tube.<br> |
| - | Incubated at 37<sup>o</sup>C for 1.5 hours. | + | Incubated at 37<sup>o</sup>C for 1.5 hours. <br> |
| - | + | [[image:Lethbridge_100823HBMaxipreps.jpg|200px]] | |
| + | ---- | ||
| + | (In Lab: AV)<br> | ||
| + | <b>Objective:</b> To PCR amplify pSB1T3, pSB1C3, pSB1A3 and run on 1% agarose gel. <br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x6.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>14.9<td>96.85 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>13 | ||
| + | <tr><td>dNTPs<td>1<td>6.5 | ||
| + | <tr><td>SB-prep-2 Primer<td>0.7<td>4.55 | ||
| + | <tr><td>SB-prep-3p Primer<td>0.7<td>4.55 | ||
| + | <tr><td>Template DNA<td>0.5<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>1.3 | ||
| + | </table><br> | ||
| + | |||
| + | Added 19.5(µL) to each tube. | ||
| + | Ran on program 6 of thermocycler. | ||
| + | |||
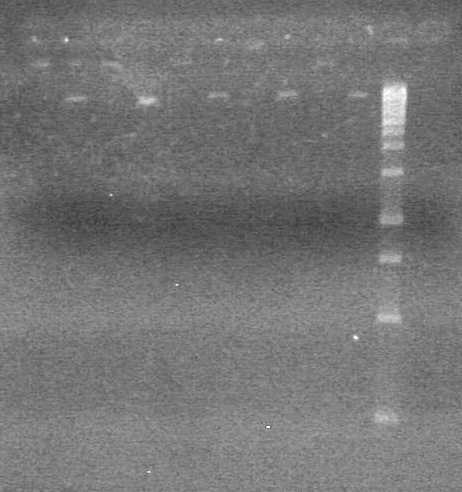
| + | <b>Results:</b> Nothing visible on gel, therefore will have to try loading a larger volume of PCR product.<br> | ||
| + | [[image:Lethbridge_100823AVBackbonePCR.jpg|100px]] | ||
| + | |||
| + | ==<font color="white">Aug 24, 2010== | ||
| + | |||
| + | (In Lab: AV)<br> | ||
| + | <b>Objective:</b> To re-run a 1% agarose gel of PCR products from Aug 23, 2010. <br> | ||
| + | <b>Results:</b> Nothing appeared on gel, therefore the PCR was unsuccessful. <br> | ||
| + | |||
| + | ==<font color="white">Aug 25, 2010== | ||
| + | |||
| + | (In Lab: JV)<br> | ||
| + | <b>Objective:</b> Create amounts of pSB1C3. <br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x6.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>14.9<td>96.85 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>13 | ||
| + | <tr><td>dNTPs<td>1<td>6.5 | ||
| + | <tr><td>SB-prep-2 Primer<td>0.7<td>4.55 | ||
| + | <tr><td>SB-prep-3p Primer<td>0.7<td>4.55 | ||
| + | <tr><td>Template DNA<td>0.5<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>1.3 | ||
| + | </table><br> | ||
| + | |||
| + | Added 19.5(µL) to each tube. | ||
| + | |||
| + | PCR- Conditions: | ||
| + | 1. 95<sup>o</sup>C for 5 min | ||
| + | 2. 94<sup>o</sup>C for 30 sec | ||
| + | 3. 55<sup>o</sup>C for 30 sec | ||
| + | 4. 68<sup>o</sup>C for 4 min | ||
| + | 5. 68<sup>o</sup>C for 10 min | ||
| + | 6. 4<sup>o</sup>C infinitely | ||
| + | (36 cycles) | ||
| + | |||
| + | ==<font color="white">Aug 25, 2010== | ||
| + | (In Lab: FM)<br> | ||
| + | <b>Objective:</b> Gradient PCR of xylE. <br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x10)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>5.8<td>58 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>1<td>10 | ||
| + | <tr><td>dNTPs<td>1<td>10 | ||
| + | <tr><td>Standard Prefix or Fusion Prefix Primer<td>0.5<td>5 | ||
| + | <tr><td>Standard Suffix or Fusion Suffix Primer<td>0.5<td>5 | ||
| + | <tr><td>Template DNA<td>1<td>10 | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>2 | ||
| + | </table><br> | ||
| + | |||
| + | Gradient Temperatures: | ||
| + | 58.5<sup>o</sup>C, 60.5<sup>o</sup>C, 62.3<sup>o</sup>C, 64.1<sup>o</sup>C, 65.9<sup>o</sup>C, 67.7<sup>o</sup>C, 69.4<sup>o</sup>C, 71.1<sup>o</sup>C | ||
| + | ----- | ||
| + | (In Lab: JS)<br> | ||
| + | <b>Objective:</b> Run a 2% agarose gel of gradient PCR of xylE. <br> | ||
| + | |||
| + | [[image:Lethbridge_100826 xylE PCR Fix.jpg|200px]] | ||
| + | |||
| + | ==<font color="white">Aug 26, 2010== | ||
| + | (In Lab: KG)<br> | ||
| + | <b>Objective:</b> Do PCR from Aug 25, 2010 to compare PCR of part from registry and our pSB1C3. <br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x2)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>14.9<td>29.8 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>4 | ||
| + | <tr><td>dNTPs<td>1<td>2 | ||
| + | <tr><td>SB-prep-26 Primer<td>0.7<td>1.4 | ||
| + | <tr><td>SB-prep-3P Primer<td>0.7<td>1.4 | ||
| + | <tr><td>Template DNA<td>0.5<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td> | ||
| + | </table><br> | ||
| + | |||
| + | PCR- Conditions: | ||
| + | 1. 95<sup>o</sup>C for 5 min | ||
| + | 2. 94<sup>o</sup>C for 30 sec | ||
| + | 3. 55<sup>o</sup>C for 30 sec | ||
| + | 4. 68<sup>o</sup>C for 4 min | ||
| + | 5. 68<sup>o</sup>C for 10 min | ||
| + | 6. 4<sup>o</sup>C infinitely | ||
| + | (36 cycles) | ||
| + | |||
| + | Ran a 1% agarose gel of gradient PCR of xylE. | ||
| + | Was stained in ethidium bromide for too long. | ||
| + | |||
| + | ==<font color="white">Aug 31, 2010== | ||
| + | (In Lab: DM)<br> | ||
| + | <b>Objective:</b> Overexpression test of xylE part (BBa_K118021) in DH5alpha cells in m9 minimal media. <br> | ||
| + | <b>Method:</b> [[Team:Lethbridge/Notebook/Protocols|Overexpression]] | ||
| + | |||
| + | 0.5 M Catechol was made, to allow for the addition of 1(µL) of this stock solution to be added to the samples taken during the overexpression. | ||
| + | Upon addition of catechol, the solutions turned yellow! | ||
| + | 1 mL samples were taken for SDS-PAGE analysis. | ||
| + | Optical density readings at 600 nm and absorbance readings at 375 and 275 nm were recorded for flasks containing either glucose or sucrose. | ||
| + | ---- | ||
| + | (In Lab: ADS)<br> | ||
| + | <b>Objective:</b> PCR amplify plasmid backbones (pSB1C3, pSB1A3 and pSB1T3). <br> | ||
| + | <b>Method:</b> Use PCR product from Aug 13, 2010 as template DNA. | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x3.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>14.4<td>52.4 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>7 | ||
| + | <tr><td>dNTPs<td>1<td>3.5 | ||
| + | <tr><td>SB-prep-26 Primer<td>0.7<td>2.45 | ||
| + | <tr><td>SB-prep-3P Primer<td>0.7<td>2.45 | ||
| + | <tr><td>Template DNA<td>1<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>0.7 | ||
| + | </table><br> | ||
| + | |||
| + | Added 19(µ:L) to each tube. | ||
| + | Phusion polymerase was used and apparently in the other previously unsuccessful PCRs, instead of Pfu polymerase. | ||
| + | |||
| + | PCR- Conditions: | ||
| + | 1. 95<sup>o</sup>C for 5 min | ||
| + | 2. 94<sup>o</sup>C for 30 sec | ||
| + | 3. 55<sup>o</sup>C for 30 sec | ||
| + | 4. 68<sup>o</sup>C for 4 min | ||
| + | 5. 68<sup>o</sup>C for 10 min | ||
| + | 6. 4<sup>o</sup>C infinitely | ||
| + | (36 cycles) | ||
| + | |||
| + | Analyzed PCR on 1% TAE agarose gel which ran for 60 min at 100 V. | ||
| + | ---- | ||
| + | <b>Objective:</b> Obtain new sources of pSB1T3, pSB1C3 and pSB1A3 plasmid backbone. Backbone from registry used up. <br> | ||
| + | |||
| + | <b>Method:</b> pSB1A3 can be obtained from anything team already possesses. pSB1C3 can be obtained from BBa_J04450 (RFP) in kit. Don't have tetracycline plates so cannot obtain pSB1T3. Used competent cell transformation protocol. <br> | ||
| + | |||
| + | <b>Method:</b> pSB1A3 can be obtained from anything team already possesses. pSB1C3 can be obtained from BBa_J04450 (RFP) in kit. Don't have tetracycline plates so cannot obtain pSB1T3. Used [[Team:Lethbridge/Notebook/Protocols|Competent Cell Transformation]] protocol. <br> | ||
| + | |||
| + | * changes: | ||
| + | **used 50µL aliquottes of DH5&alpha | ||
| + | **did not pipette up and down once, the cells were just swirled 3 times | ||
| + | **added 500µL SOC media, shoock at 37<sup>0</sup>C for 90 min | ||
| + | **platted 250µL and 150µL<br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <td><b>Results</b> | ||
| + | <tr><td>contents<td><b>&250µL</b><td><b>150µL</b> | ||
| + | <tr><td>+ control(pUC19)<td>good<td>good | ||
| + | <tr><td>J04450<td>X (TMTC)<td>X(TMTC) | ||
| + | </table><br> | ||
| + | |||
| + | Prepared overnight cultures for minipreps. Added 5(µL) of 35 mg/mL Chloramphenicl to 5 mL of LB Media. Inoculated media with cells from single colony picked from transformation plate. Incubated overnight at 37<sup>o</sup>C with shaking. | ||
| + | <br><br> | ||
 "
"