Team:Lethbridge/Notebook/Lab Work/August
From 2010.igem.org
Adam.smith4 (Talk | contribs) (→Aug 10, 2010) |
Liszabruder (Talk | contribs) |
||
| (18 intermediate revisions not shown) | |||
| Line 862: | Line 862: | ||
</table><br> | </table><br> | ||
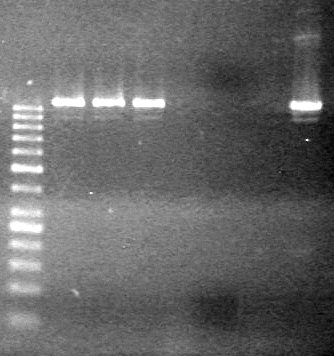
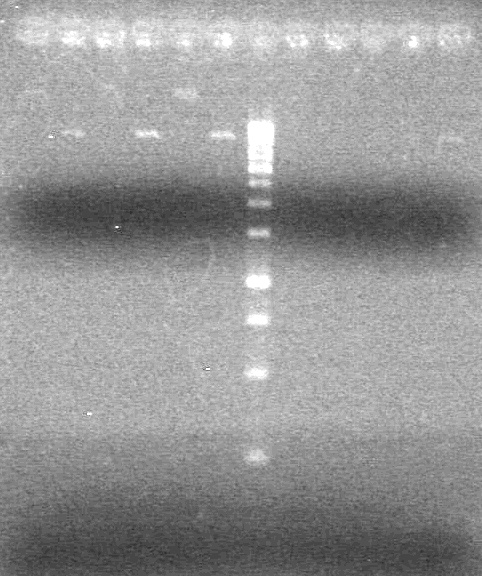
| - | Ran samples on 1.5% agarose gel (1X TAE) for 60 minutes at 100V. | + | Ran samples on 1.5% agarose gel (1X TAE) for 60 minutes at 100V. <br> |
| - | + | <b>Results:</b><br> | |
| + | [[image:Lethbridge_100810xylE PCR AS.JPG|100px]] | ||
==<font color="white">Aug 11, 2010== | ==<font color="white">Aug 11, 2010== | ||
| Line 922: | Line 923: | ||
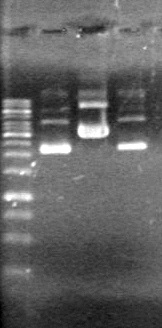
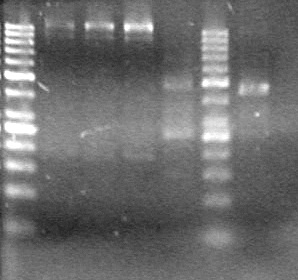
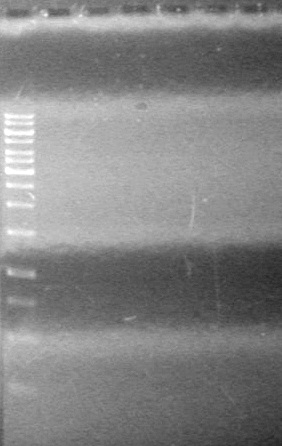
<b>Results:</b> Lanes 2, 3, 4 and 8 showed PCR amplification. Colonies chosen don't show the correct insert size. <br> | <b>Results:</b> Lanes 2, 3, 4 and 8 showed PCR amplification. Colonies chosen don't show the correct insert size. <br> | ||
| + | [[image:Lethbridge_100812 JV AS Colony PCR Pfu.JPG|200px]] | ||
---- | ---- | ||
| Line 950: | Line 952: | ||
</table><br> | </table><br> | ||
| - | Added 18µL Master Mix to each reaction tube. | + | Added 18µL Master Mix to each reaction tube.<br> |
| - | Analyzed PCR products on 2.5% TAE gel. | + | Analyzed PCR products on 2.5% TAE gel.<br> |
| - | + | <b>Results:</b><br> | |
| + | [[image:Lethbridge_100812ASJVMassColonyPCRLargeGel (1).JPG|200px]]<br><br> | ||
| + | [[image:Lethbridge_100812ASJVMassColonyPCRSmallGel.JPG|200px]] | ||
---- | ---- | ||
| Line 981: | Line 985: | ||
</table><br> | </table><br> | ||
| - | Added 18µL Master Mix to each reaction tube. | + | Added 18µL Master Mix to each reaction tube.<br> |
| - | Analyzed PCR products on 2.5% TAE gel run at 100 V for 35 minutes. | + | Analyzed PCR products on 2.5% TAE gel run at 100 V for 35 minutes.<br> |
| - | + | [[image:Lethbridge_100813ADS Awesome PCR.JPG|200px]] | |
==<font color="white">Aug 13, 2010== | ==<font color="white">Aug 13, 2010== | ||
| Line 1,003: | Line 1,007: | ||
</table><br> | </table><br> | ||
| - | Added 49µL Master Mix to each reaction tube. | + | Added 49µL Master Mix to each reaction tube.<br> |
| + | |||
---- | ---- | ||
| Line 1,030: | Line 1,035: | ||
(36 cycles) | (36 cycles) | ||
| - | Added 19.5µL Master Mix to each reaction tube. | + | Added 19.5µL Master Mix to each reaction tube.<br> |
| - | Analyzed products on 1% TAE agarose gel which ran for 60 minutes at 100 V. | + | Analyzed products on 1% TAE agarose gel which ran for 60 minutes at 100 V.<br> |
| - | + | <b>Results:</b><br> | |
| + | [[image:Lethbridge_100814PlasmidBackbones.JPG|100px]] | ||
==<font color="white">Aug 14, 2010== | ==<font color="white">Aug 14, 2010== | ||
| Line 1,062: | Line 1,068: | ||
<tr><td>19<td>MT | <tr><td>19<td>MT | ||
<tr><td>20<td>MT | <tr><td>20<td>MT | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</table> | </table> | ||
| Line 1,102: | Line 1,101: | ||
</table> | </table> | ||
| - | + | [[image:Lethbridge_100815MiniprepPCR.JPG|200px]] | |
---- | ---- | ||
| Line 1,134: | Line 1,133: | ||
Incubated at 37<sup>o</sup>C. | Incubated at 37<sup>o</sup>C. | ||
| - | Analyzed results on 2.5% TAE agarose gel which ran at 100 V for 50 minutes. | + | Analyzed results on 2.5% TAE agarose gel which ran at 100 V for 50 minutes.<br> |
| - | + | [[image:Lethbridge_100814ColonyPCRRetryandStartofassembly.JPG|200px]] | |
<b>Results:</b> Lost all the DNA in the column clean-up step and will have to re-do.<br> | <b>Results:</b> Lost all the DNA in the column clean-up step and will have to re-do.<br> | ||
| Line 1,145: | Line 1,144: | ||
<b>Objective:</b> Assemble mms6-dT and lumazine-dT using three antibiotic assembly. <br> | <b>Objective:</b> Assemble mms6-dT and lumazine-dT using three antibiotic assembly. <br> | ||
| - | <b>Method:</b> | + | <b>Method:</b> |
| + | #PCR amplify BioBricks (Prefix/Suffix) | ||
| + | #Restrict BioBricks | ||
| + | #Ligate BioBricks into psB1C3 | ||
| + | #Confirm ligation by PCR analysis (VF2/VR) | ||
| + | #Transform ligation mixes | ||
| + | #Screen colonies with Colony PCR | ||
<u>PCR:</u> Thermocycler set to iGEM program 11<br> | <u>PCR:</u> Thermocycler set to iGEM program 11<br> | ||
| Line 1,185: | Line 1,190: | ||
</table> | </table> | ||
| - | For | + | For pSB1C3 - |
<table><table border ="3"> | <table><table border ="3"> | ||
<tr><td><b>Ingredient</b><td><b>Reaction Mix(µL)</b> | <tr><td><b>Ingredient</b><td><b>Reaction Mix(µL)</b> | ||
| Line 1,273: | Line 1,278: | ||
</table><br> | </table><br> | ||
| - | Added 15 (µL) MM to each tube. | + | Added 15 (µL) MM to each tube.<br> |
| + | <b>Results:</b><br> | ||
==<font color="white">Aug 15, 2010== | ==<font color="white">Aug 15, 2010== | ||
| Line 1,378: | Line 1,384: | ||
Added 45 (µL) MM to each tube. | Added 45 (µL) MM to each tube. | ||
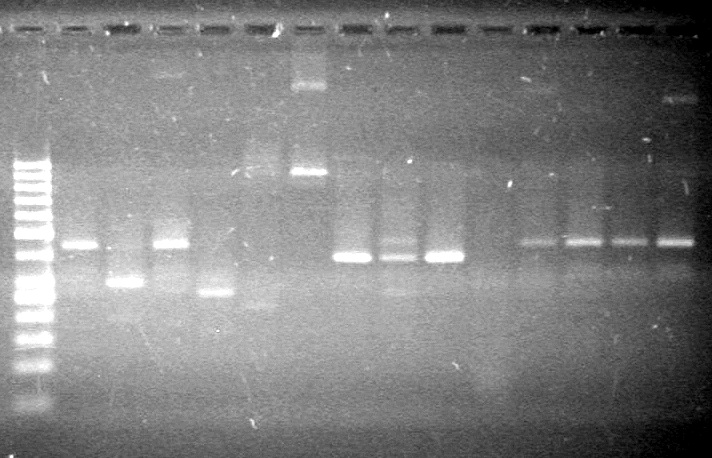
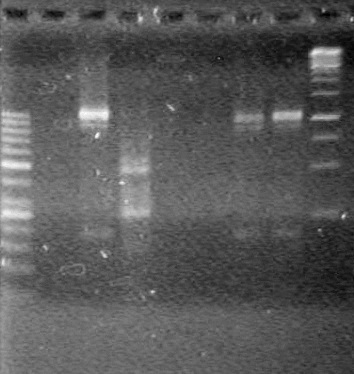
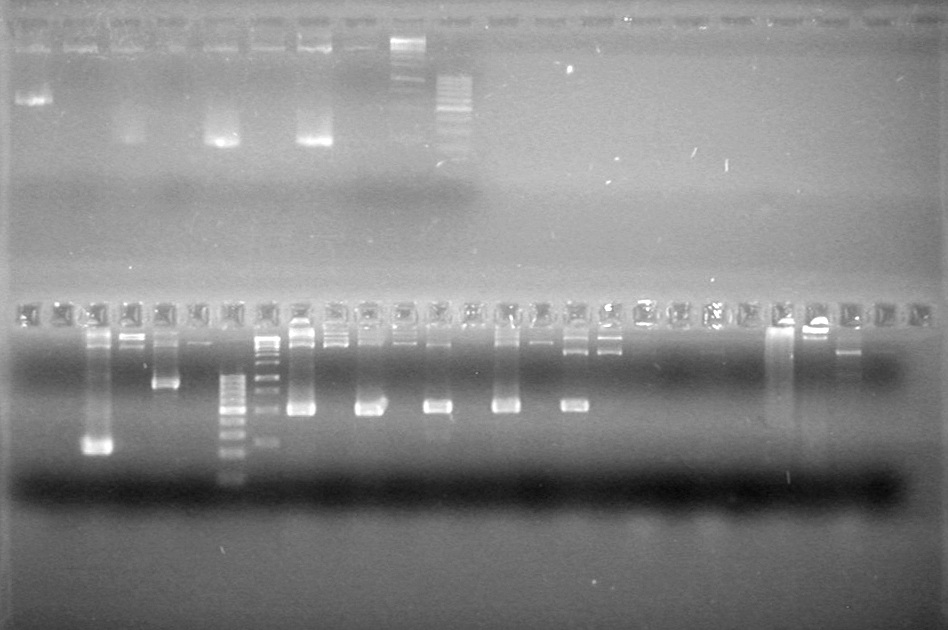
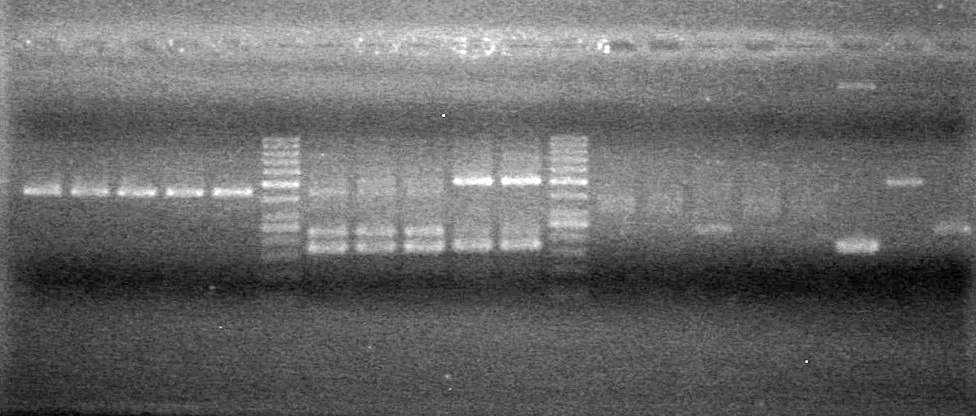
| - | Analyzed PCR products of BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. | + | Analyzed PCR products of BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. <br> |
| + | <b>Results:</b><br> | ||
| + | [[image:Lethbridge_100815PCRAssembly.JPG|200px]] | ||
| + | |||
| - | |||
---- | ---- | ||
(In Lab: ADS)<br> | (In Lab: ADS)<br> | ||
| Line 1,445: | Line 1,453: | ||
</table><br> | </table><br> | ||
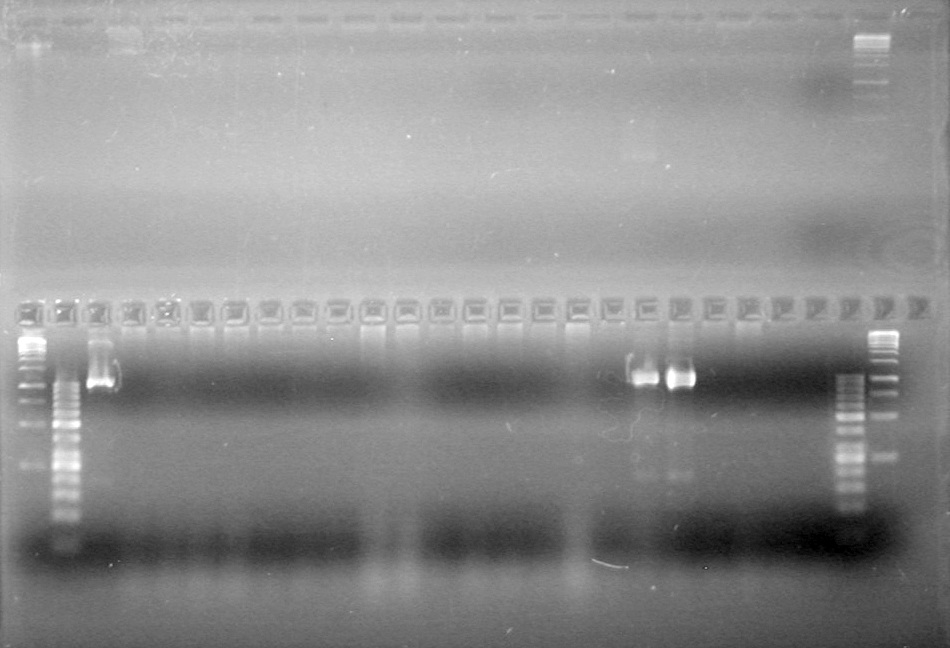
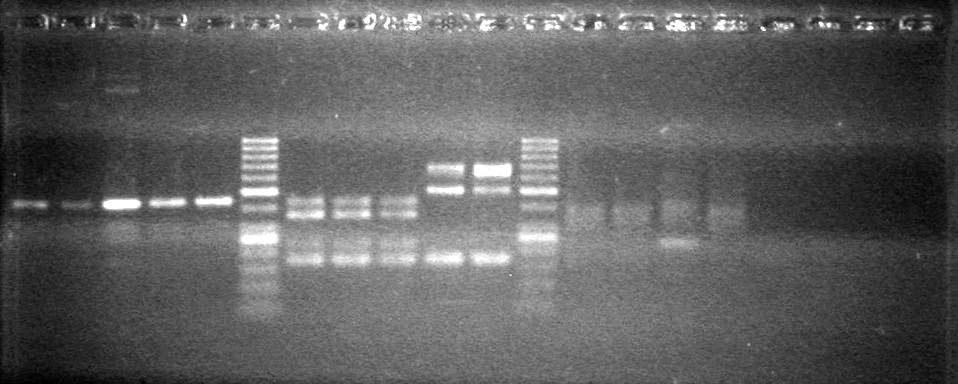
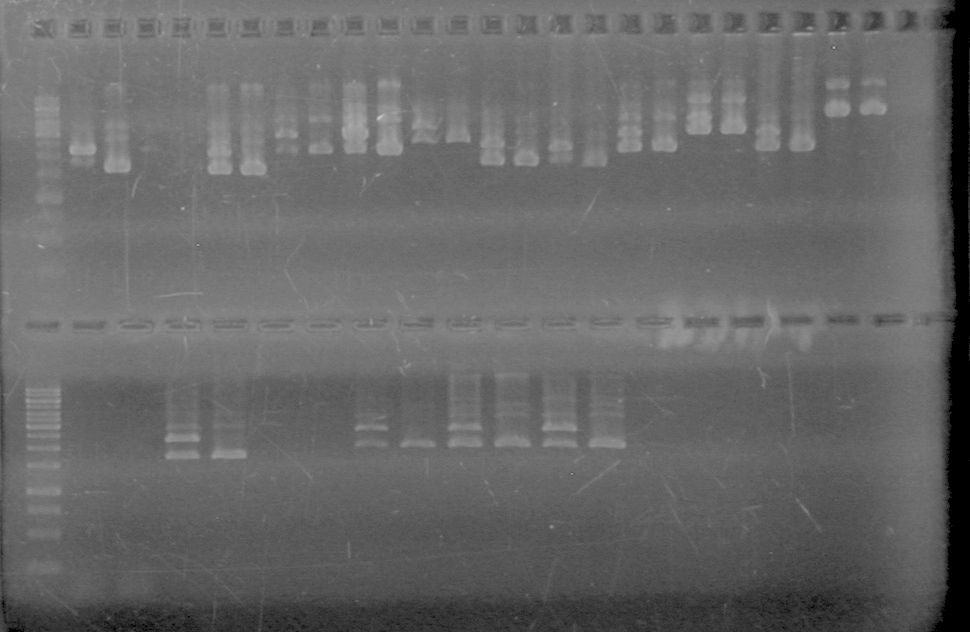
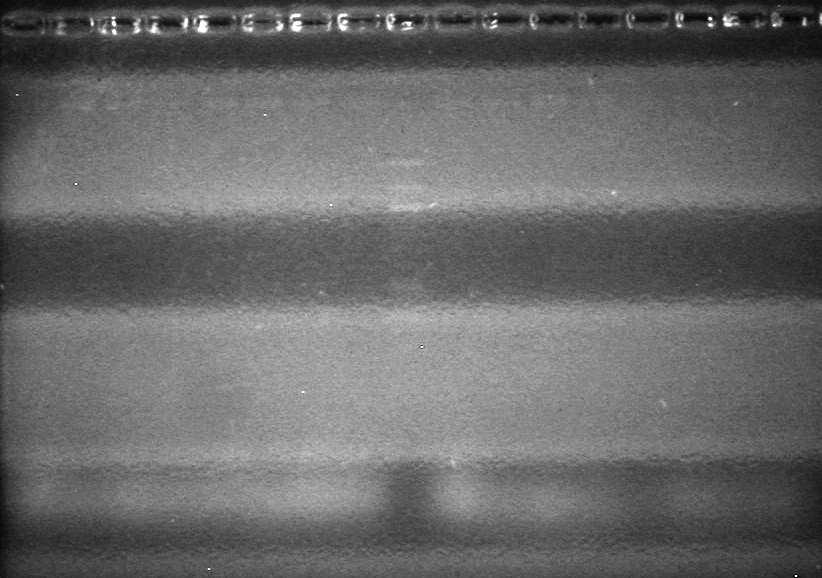
| - | Analyzed PCR products of overnight BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. | + | Analyzed PCR products of overnight BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. <br> |
| - | + | <b>Results:</b><br> | |
| - | + | [[image:Lethbridge_100816PostLigationPCRScreen.JPG|200px]] | |
==<font color="white">Aug 16, 2010 Evening == | ==<font color="white">Aug 16, 2010 Evening == | ||
| Line 1,523: | Line 1,531: | ||
</table><br> | </table><br> | ||
| - | Ran a 2% Agarose gel in 1X TAE buffer for 65 minutes at 100V. | + | Ran a 2% Agarose gel in 1X TAE buffer for 65 minutes at 100V.<br> |
| - | + | [[image:Lethbridge_100817PostLigationPCRTest.JPG|100px]] | |
| - | + | ||
==<font color="white">Aug 17, 2010 Evening == | ==<font color="white">Aug 17, 2010 Evening == | ||
| Line 1,650: | Line 1,657: | ||
Ran on cycle 4 of thermocycler. | Ran on cycle 4 of thermocycler. | ||
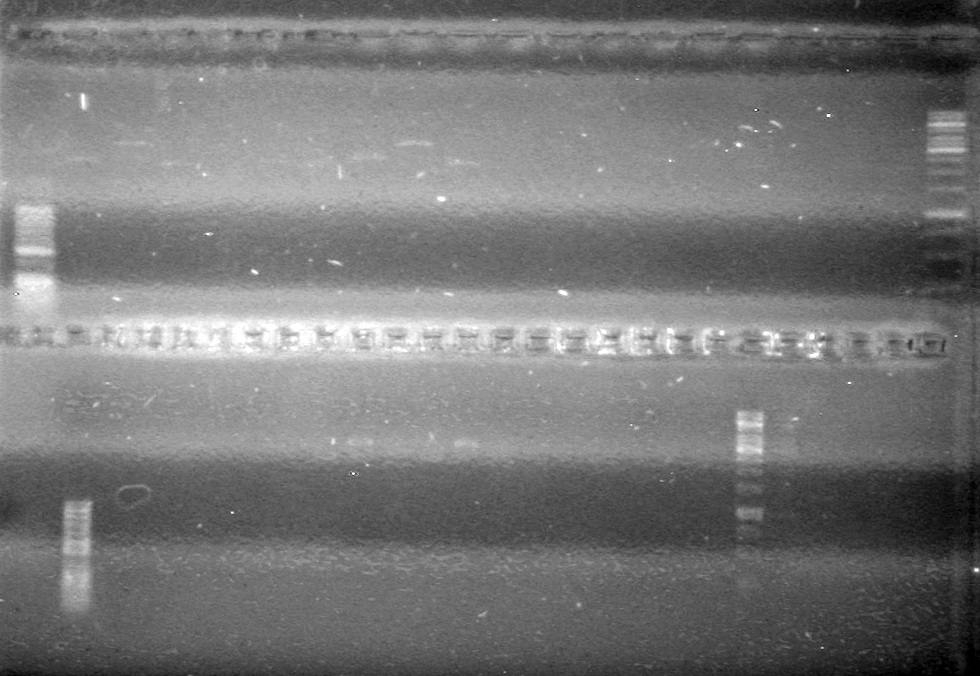
| - | Viewed PCRs on 2% TAE agarose gel that ran at 110 V for 30 minutes. | + | Viewed PCRs on 2% TAE agarose gel that ran at 110 V for 30 minutes.<br> |
| - | + | [[image:Lethbridge_100818AssemblyandHeatKillPCR.JPG|200px]] | |
| - | + | ||
---- | ---- | ||
(In Lab: HB)<br> | (In Lab: HB)<br> | ||
| Line 1,744: | Line 1,750: | ||
Samples were run on a 2% agarose gel in 1X TAE Buffer. | Samples were run on a 2% agarose gel in 1X TAE Buffer. | ||
| - | |||
| - | |||
---- | ---- | ||
| Line 1,829: | Line 1,833: | ||
---- | ---- | ||
(In Lab: JV)<br> | (In Lab: JV)<br> | ||
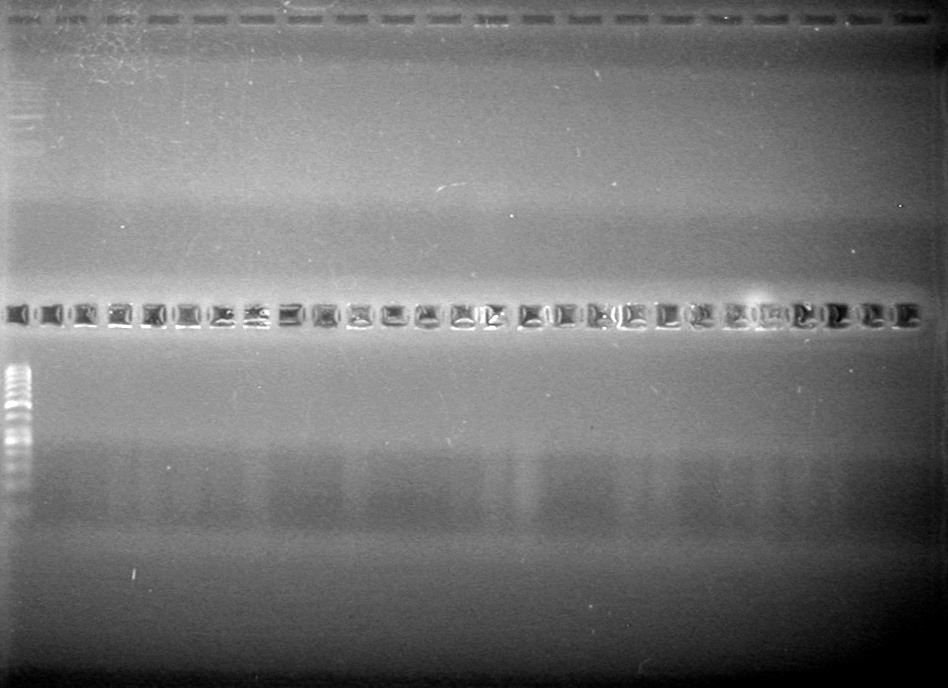
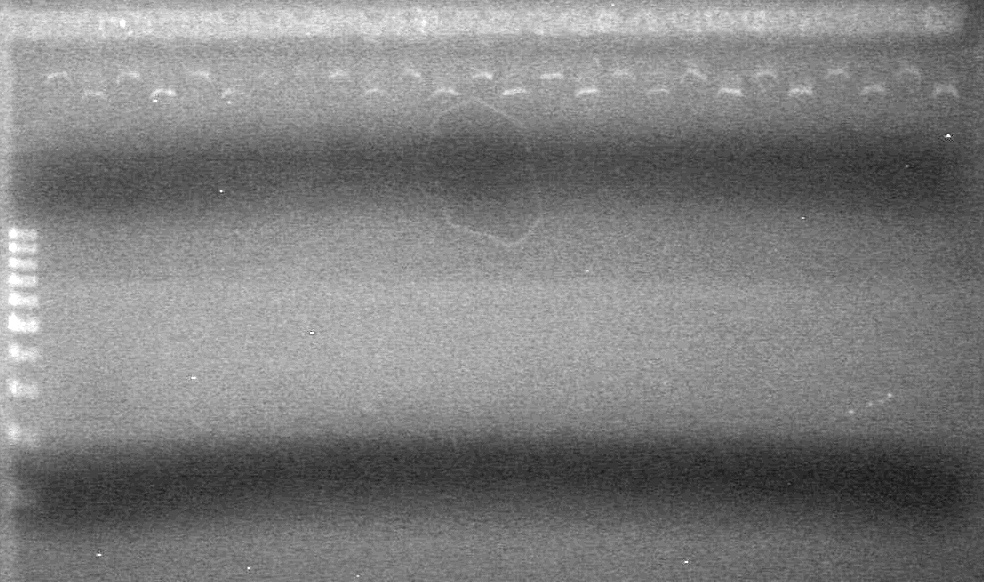
| - | Analyzed HB's PCR products on 2% TAE gel which ran for 60 minutes at 100 V. | + | Analyzed HB's PCR products on 2% TAE gel which ran for 60 minutes at 100 V.<br> |
| - | + | [[image:Lethbridge_100819PCRMania.JPG|200px]] | |
| - | + | ||
==<font color="white">Aug 20, 2010== | ==<font color="white">Aug 20, 2010== | ||
| Line 1,843: | Line 1,846: | ||
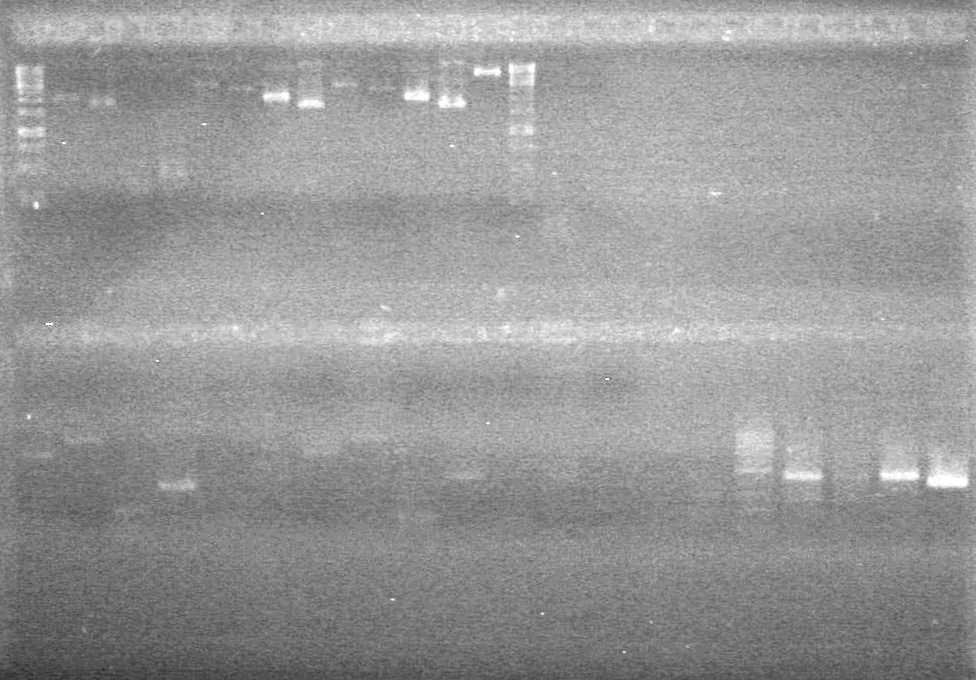
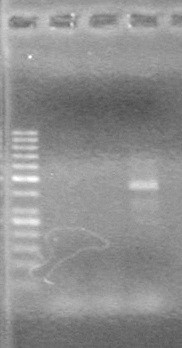
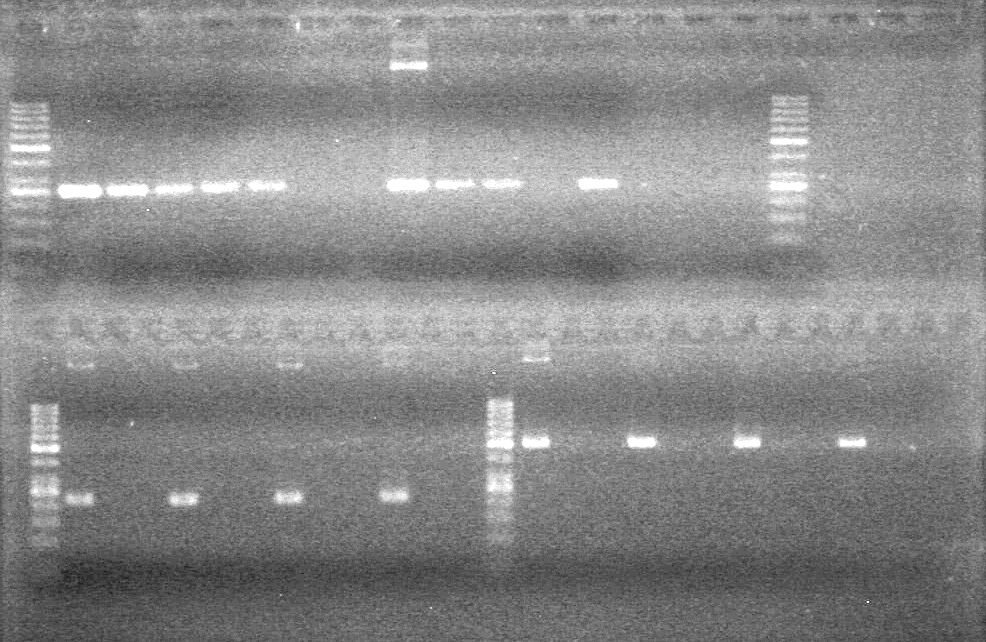
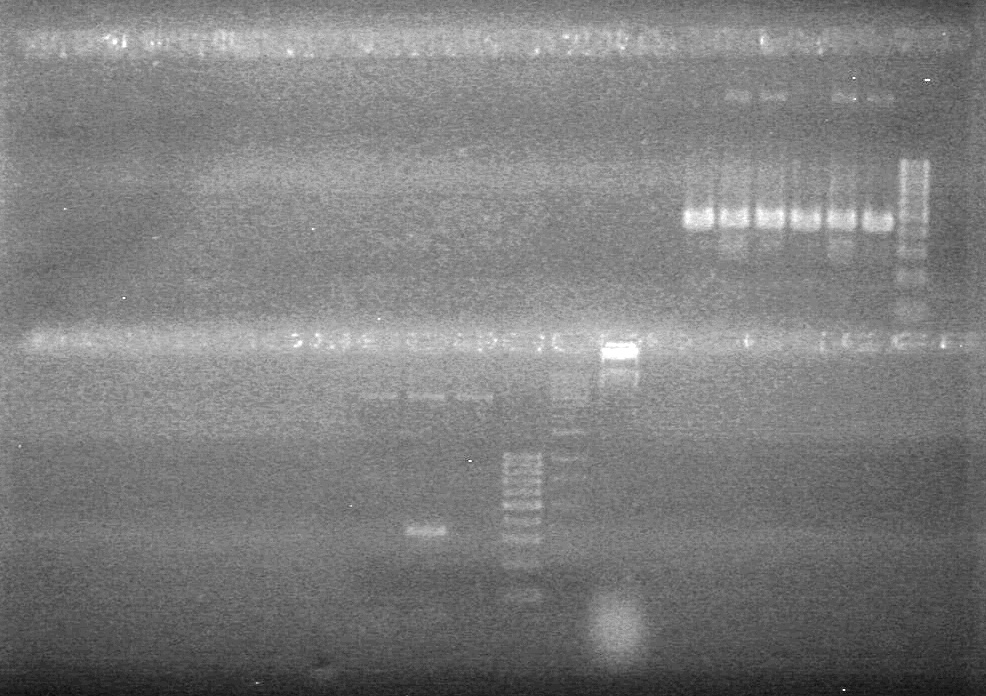
<b>Results:</b> DNA concentration was good, however there was no evidence of an insert into pET-28(A). <br> | <b>Results:</b> DNA concentration was good, however there was no evidence of an insert into pET-28(A). <br> | ||
| - | + | [[image:Lethbridge_100819pET28aTransformScreenGel1.JPG|200px]]<br><br> | |
| + | [[image:Lethbridge_100819pET28aTransformScreenGel2.JPG|200px]]<br><br> | ||
| + | [[image:Lethbridge_100819pET28aTransformScreenGel3.JPG|200px]]<br><br> | ||
| + | [[image:Lethbridge_100819pET28aTransformScreenGel4.JPG|200px]]<br><br> | ||
==<font color="white">Aug 23, 2010== | ==<font color="white">Aug 23, 2010== | ||
| Line 1,879: | Line 1,885: | ||
</table> | </table> | ||
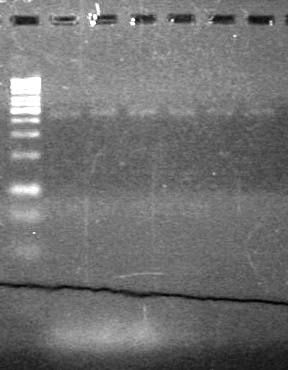
| - | 15(µL) added to each rxn tube. | + | 15(µL) added to each rxn tube.<br> |
| - | Incubated at 37<sup>o</sup>C for 1.5 hours. | + | Incubated at 37<sup>o</sup>C for 1.5 hours. <br> |
| - | + | [[image:Lethbridge_100823HBMaxipreps.jpg|200px]] | |
---- | ---- | ||
(In Lab: AV)<br> | (In Lab: AV)<br> | ||
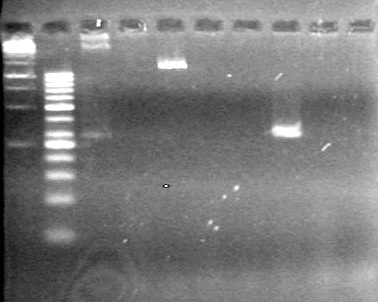
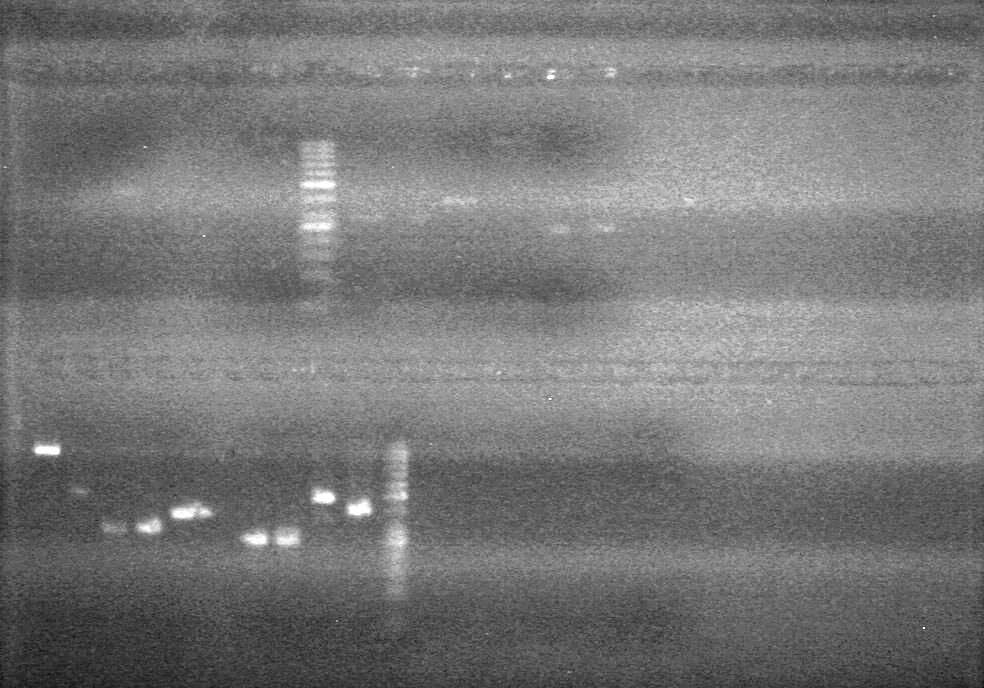
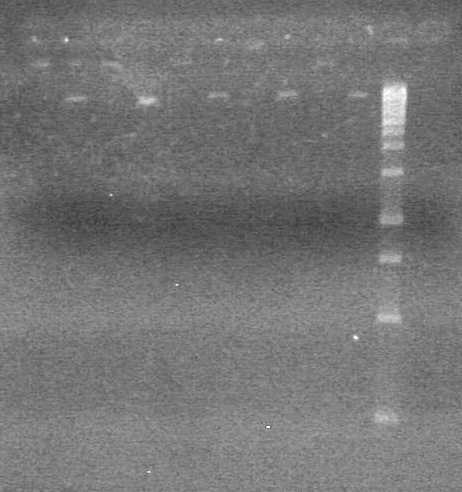
| - | <b>Objective:</b> To PCR amplify | + | <b>Objective:</b> To PCR amplify pSB1T3, pSB1C3, pSB1A3 and run on 1% agarose gel. <br> |
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 1,903: | Line 1,909: | ||
<b>Results:</b> Nothing visible on gel, therefore will have to try loading a larger volume of PCR product.<br> | <b>Results:</b> Nothing visible on gel, therefore will have to try loading a larger volume of PCR product.<br> | ||
| + | [[image:Lethbridge_100823AVBackbonePCR.jpg|100px]] | ||
==<font color="white">Aug 24, 2010== | ==<font color="white">Aug 24, 2010== | ||
| Line 1,960: | Line 1,967: | ||
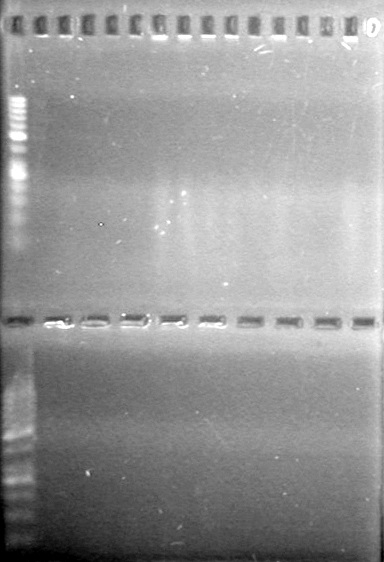
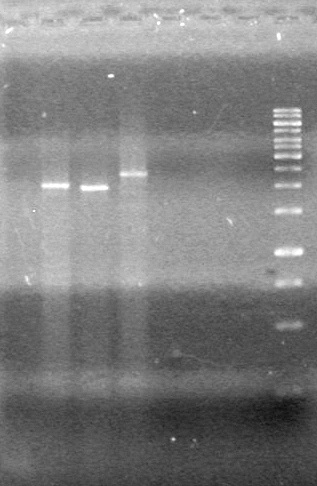
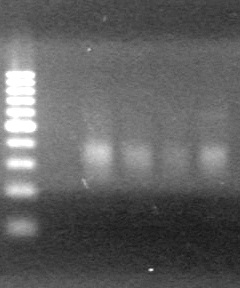
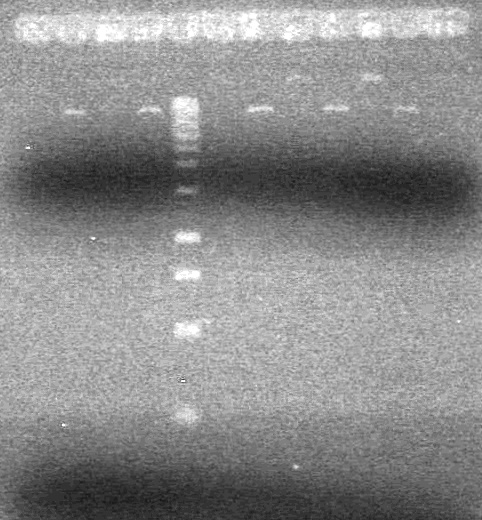
<b>Objective:</b> Run a 2% agarose gel of gradient PCR of xylE. <br> | <b>Objective:</b> Run a 2% agarose gel of gradient PCR of xylE. <br> | ||
| - | + | [[image:Lethbridge_100826 xylE PCR Fix.jpg|200px]] | |
==<font color="white">Aug 26, 2010== | ==<font color="white">Aug 26, 2010== | ||
| Line 2,049: | Line 2,056: | ||
Prepared overnight cultures for minipreps. Added 5(µL) of 35 mg/mL Chloramphenicl to 5 mL of LB Media. Inoculated media with cells from single colony picked from transformation plate. Incubated overnight at 37<sup>o</sup>C with shaking. | Prepared overnight cultures for minipreps. Added 5(µL) of 35 mg/mL Chloramphenicl to 5 mL of LB Media. Inoculated media with cells from single colony picked from transformation plate. Incubated overnight at 37<sup>o</sup>C with shaking. | ||
| + | <br><br> | ||
 "
"