Team:Lethbridge/Notebook/Lab Work/August
From 2010.igem.org
(→Aug 26, 2010) |
Liszabruder (Talk | contribs) |
||
| (33 intermediate revisions not shown) | |||
| Line 209: | Line 209: | ||
<b>Method:</b> Used [[Team:Lethbridge/Notebook/Protocols|Restriction of Plasmid DNA]] protocol. | <b>Method:</b> Used [[Team:Lethbridge/Notebook/Protocols|Restriction of Plasmid DNA]] protocol. | ||
| - | * A front | + | * A front vector was made made in sRBS ,mRBS, dT plasmids using EcoRI and Xbal enzymes |
* pDNA that was cut out of plasmid for a front vector was pBAD, TetR, dT and pLacI using EcoRI and SpeI enzymes | * pDNA that was cut out of plasmid for a front vector was pBAD, TetR, dT and pLacI using EcoRI and SpeI enzymes | ||
* A back vector was made in sRBS and mRBS plasmids with PstI and SpeI | * A back vector was made in sRBS and mRBS plasmids with PstI and SpeI | ||
| Line 252: | Line 252: | ||
</table> | </table> | ||
*Added 48.8 µL of Master Mix to each PCR reaction<br><br><br> | *Added 48.8 µL of Master Mix to each PCR reaction<br><br><br> | ||
| + | |||
| + | |||
<b>Objective:</b> Complete maxipreps of Lumazine (K249002), EYFP (E0030), and ECFP (E0020) and run on 1% agarose gel.<br> | <b>Objective:</b> Complete maxipreps of Lumazine (K249002), EYFP (E0030), and ECFP (E0020) and run on 1% agarose gel.<br> | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 262: | Line 264: | ||
</table> | </table> | ||
*Ran at 100V for 42 minutes. Stained in EtBr for 15 minutes.<br> | *Ran at 100V for 42 minutes. Stained in EtBr for 15 minutes.<br> | ||
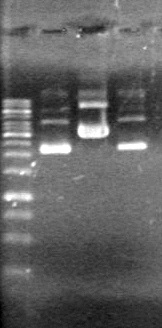
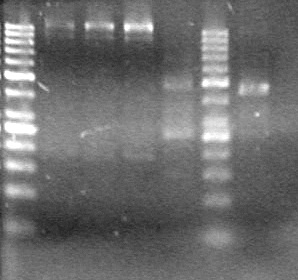
| - | <b>Results:</b> | + | <b>Results:</b> [[image:Lethbridge_100803JV Maxiprep.JPG|50px]]<br><br><br> |
| + | |||
| + | |||
<b>Objective:</b> Ligate dT into pSB1C3.<br> | <b>Objective:</b> Ligate dT into pSB1C3.<br> | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 331: | Line 335: | ||
<tr><td>20<td>pLacI PCR product<td> good | <tr><td>20<td>pLacI PCR product<td> good | ||
</table><br> | </table><br> | ||
| + | |||
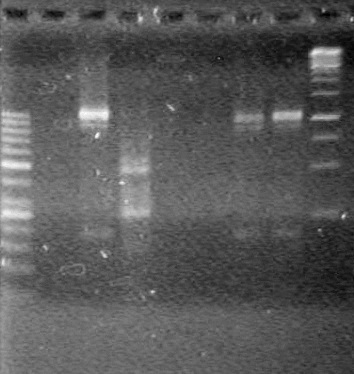
| + | [[image:Lethbridge_100803AS.JPG|200px]] | ||
---- | ---- | ||
<b>Objective:</b> To ligate: pBAD-sRBS/mRBS, sRBS/mRBS-TetR, TetR-dT, dT-pTet, pLacI-sRBS/mRBS, Mms6-ptet28(a), dT-PSB1C3<br> | <b>Objective:</b> To ligate: pBAD-sRBS/mRBS, sRBS/mRBS-TetR, TetR-dT, dT-pTet, pLacI-sRBS/mRBS, Mms6-ptet28(a), dT-PSB1C3<br> | ||
| - | <b>Method:</b> [[Team:Lethbridge/Notebook/Protocols|Ligation of Plasmid DNA]] | + | <b>Method:</b> [[Team:Lethbridge/Notebook/Protocols|Ligation of Plasmid DNA]]<br> |
| - | + | 15µL pDNA in plasmid, and 15 µL of pDNA biobrick | |
==<font color="white">August 4, 2010== | ==<font color="white">August 4, 2010== | ||
| Line 392: | Line 398: | ||
*Ran at 100V for 70 minutes. | *Ran at 100V for 70 minutes. | ||
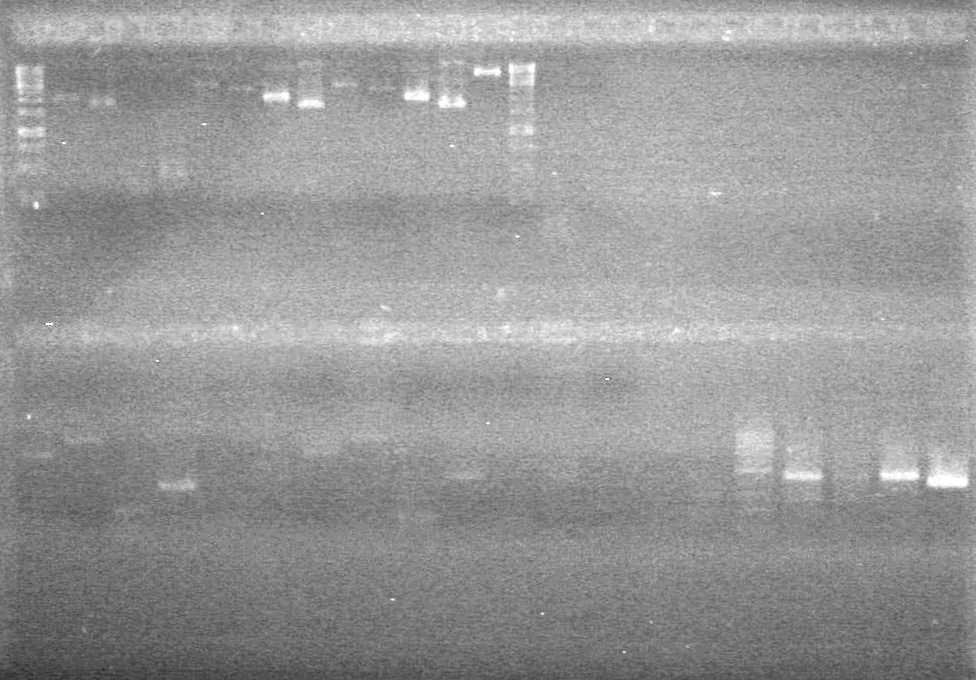
| - | <b>Results:</b> | + | <b>Results:</b>[[image:Lethbridge_100804 JV Ligation PCR AS.JPG|200px]] <br><br><br> |
| + | |||
| + | |||
| + | |||
<b>Objective:</b> Transform the successful ligations<br> | <b>Objective:</b> Transform the successful ligations<br> | ||
| Line 454: | Line 463: | ||
Gel ran for 60 minutes at 100 V. Small & faint band was slightly visible. | Gel ran for 60 minutes at 100 V. Small & faint band was slightly visible. | ||
| - | Gel extraction was carried out using QIAGEN method. Eluted to 12 (µL). | + | Gel extraction was carried out using QIAGEN method. Eluted to 12 (µL).<br> |
| + | [[image:Lethbridge_100809 JV Mms6 Gel Extract BW.jpg|200px]] | ||
==<font color="white">August 6, 2010 Evening== | ==<font color="white">August 6, 2010 Evening== | ||
| Line 522: | Line 532: | ||
<tr><td>7<td>lumazine(justin's)<td>5(µL) sample, 1(µL)dye | <tr><td>7<td>lumazine(justin's)<td>5(µL) sample, 1(µL)dye | ||
</table><br> | </table><br> | ||
| + | [[image:Lethbridge_100806ADSPCR.JPG|200px]] | ||
Repeat gel with template controls | Repeat gel with template controls | ||
| Line 537: | Line 548: | ||
<tr><td>1<td>1Kb ladder<td>0.5(µL) ladder, 2(µL) dye, 9.5(µL) H<sub>2</sub>0 | <tr><td>1<td>1Kb ladder<td>0.5(µL) ladder, 2(µL) dye, 9.5(µL) H<sub>2</sub>0 | ||
</table><br> | </table><br> | ||
| + | [[image:Lethbridge_100809AS-PCR.jpg|200px]] | ||
*ran at 100V for 75 minutes | *ran at 100V for 75 minutes | ||
| Line 570: | Line 582: | ||
</table><br> | </table><br> | ||
| - | Ran at 100 V for 45 minutes. mRBS-xylE did not amplify while lumazine did amplify. | + | Ran at 100 V for 45 minutes. mRBS-xylE did not amplify while lumazine did amplify. <br> |
| - | + | <b>Results:</b> Ligation of mRBS-xylE NOT confirmed<br> | |
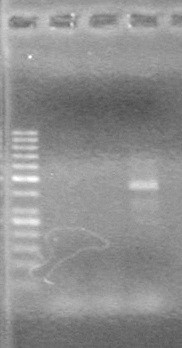
| + | [[image:Lethbridge_100809 HB xylE Lum PCR BW.jpg|50px]] | ||
| + | |||
---- | ---- | ||
| Line 656: | Line 670: | ||
<tr><td>20<td>pLacI-sRBS 3 URD | <tr><td>20<td>pLacI-sRBS 3 URD | ||
</table><br> | </table><br> | ||
| + | [[image:Lethbridge_100809AVKG Top.jpg|200px]] | ||
<table border ="3"> | <table border ="3"> | ||
| Line 681: | Line 696: | ||
</table><br> | </table><br> | ||
| - | + | [[image:Lethbridge_100809AVKG Bottom.jpg|200px]] | |
---- | ---- | ||
| Line 690: | Line 705: | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| - | + | ||
| - | + | <u>Restrictions</u><br> | |
*Restrict Lumazine wit EcoRI and SpeI (Red Buffer)<br> | *Restrict Lumazine wit EcoRI and SpeI (Red Buffer)<br> | ||
*Restrict the dT with XbaI and EcoRI (Orange Buffer)<br> | *Restrict the dT with XbaI and EcoRI (Orange Buffer)<br> | ||
| Line 706: | Line 721: | ||
Incubated reactions for 60 minutes at 37<sup>o</sup>C<br> | Incubated reactions for 60 minutes at 37<sup>o</sup>C<br> | ||
| - | + | <u>Ligation</u><br> | |
Reaction set up as follows: | Reaction set up as follows: | ||
*T4 DNA ligase - 0.25µL<br> | *T4 DNA ligase - 0.25µL<br> | ||
| Line 771: | Line 786: | ||
</table><br> | </table><br> | ||
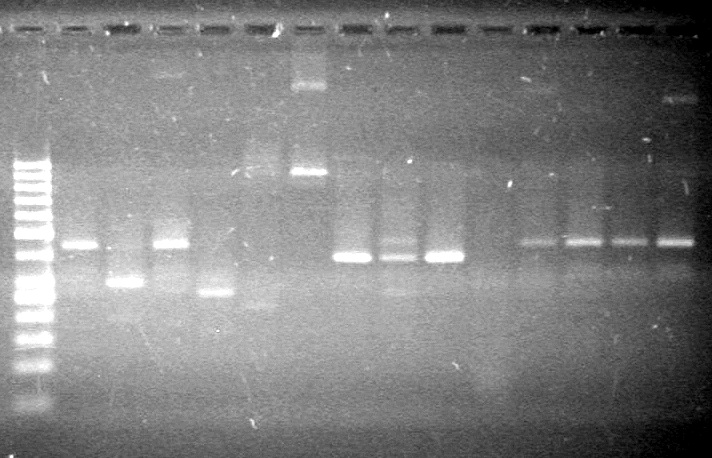
| - | + | [[image:Lethbridge_100810ReRunRestrictionDigers.JPG|200px]] | |
---- | ---- | ||
| Line 814: | Line 829: | ||
</table><br> | </table><br> | ||
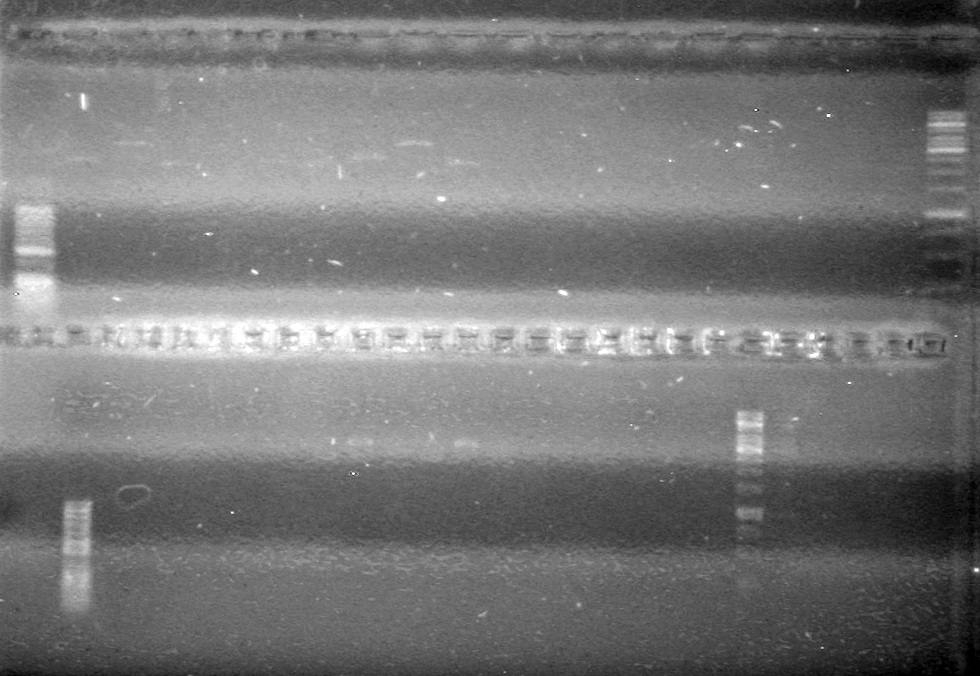
| - | Controls: sRBS, pTET & mRBS - will PCR these to compare size using same conditions as above. | + | Controls: sRBS, pTET & mRBS - will PCR these to compare size using same conditions as above. <br> |
| - | + | [[image:Lethbridge_100810MassiveColonyPCR JV AS.JPG|200px]]<br> | |
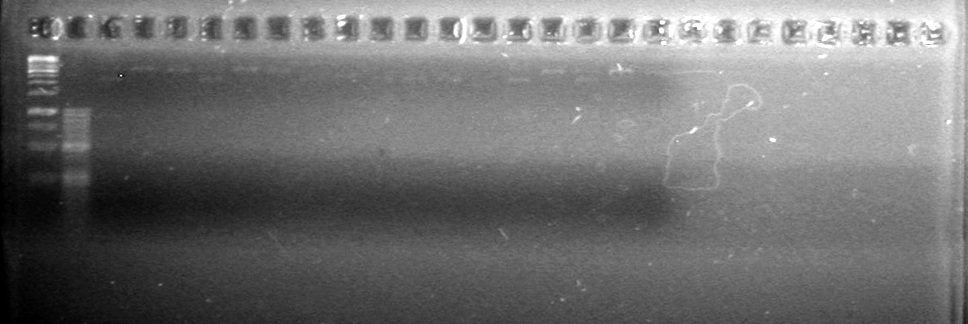
| - | + | [[image:Lethbridge_100810MassiveColonyPCR Large Gel JV AS.JPG|200px]] | |
==<font color="white">Aug 10, 2010 Evening== | ==<font color="white">Aug 10, 2010 Evening== | ||
| Line 847: | Line 862: | ||
</table><br> | </table><br> | ||
| - | Ran samples on 1.5% agarose gel (1X TAE) for 60 minutes at 100V. | + | Ran samples on 1.5% agarose gel (1X TAE) for 60 minutes at 100V. <br> |
| - | + | <b>Results:</b><br> | |
| + | [[image:Lethbridge_100810xylE PCR AS.JPG|100px]] | ||
==<font color="white">Aug 11, 2010== | ==<font color="white">Aug 11, 2010== | ||
| Line 907: | Line 923: | ||
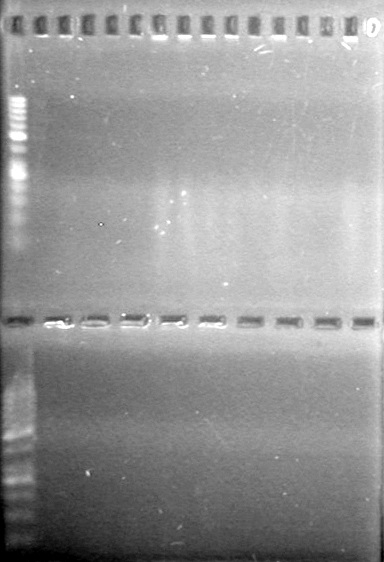
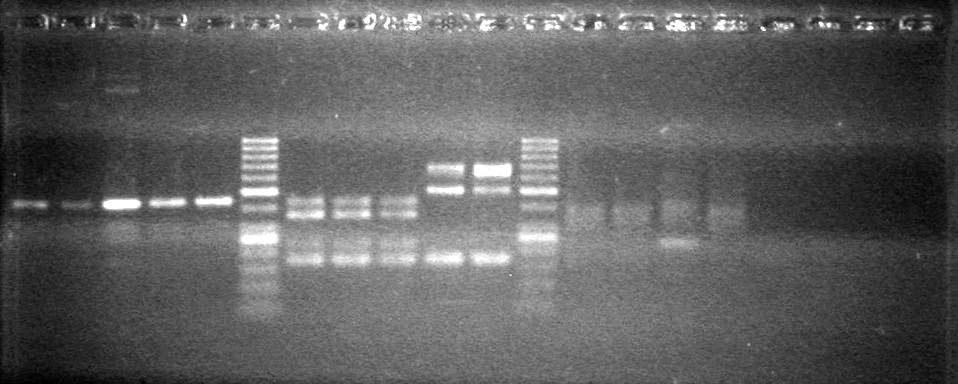
<b>Results:</b> Lanes 2, 3, 4 and 8 showed PCR amplification. Colonies chosen don't show the correct insert size. <br> | <b>Results:</b> Lanes 2, 3, 4 and 8 showed PCR amplification. Colonies chosen don't show the correct insert size. <br> | ||
| + | [[image:Lethbridge_100812 JV AS Colony PCR Pfu.JPG|200px]] | ||
---- | ---- | ||
| Line 935: | Line 952: | ||
</table><br> | </table><br> | ||
| - | Added 18µL Master Mix to each reaction tube. | + | Added 18µL Master Mix to each reaction tube.<br> |
| - | Analyzed PCR products on 2.5% TAE gel. | + | Analyzed PCR products on 2.5% TAE gel.<br> |
| - | + | <b>Results:</b><br> | |
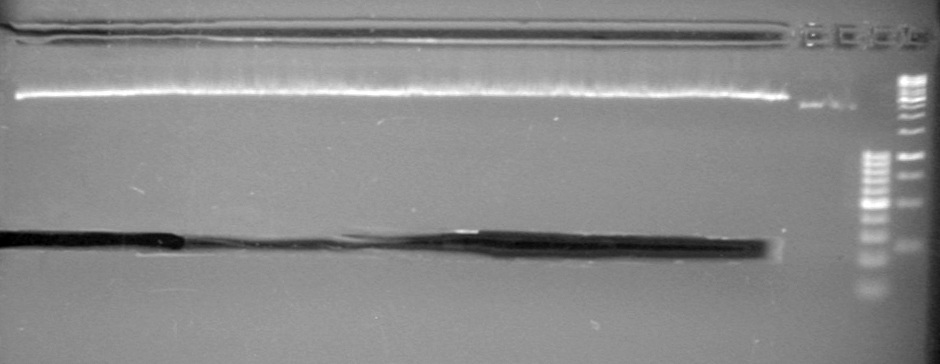
| + | [[image:Lethbridge_100812ASJVMassColonyPCRLargeGel (1).JPG|200px]]<br><br> | ||
| + | [[image:Lethbridge_100812ASJVMassColonyPCRSmallGel.JPG|200px]] | ||
---- | ---- | ||
| Line 966: | Line 985: | ||
</table><br> | </table><br> | ||
| - | Added 18µL Master Mix to each reaction tube. | + | Added 18µL Master Mix to each reaction tube.<br> |
| - | Analyzed PCR products on 2.5% TAE gel run at 100 V for 35 minutes. | + | Analyzed PCR products on 2.5% TAE gel run at 100 V for 35 minutes.<br> |
| - | + | [[image:Lethbridge_100813ADS Awesome PCR.JPG|200px]] | |
==<font color="white">Aug 13, 2010== | ==<font color="white">Aug 13, 2010== | ||
| Line 988: | Line 1,007: | ||
</table><br> | </table><br> | ||
| - | Added 49µL Master Mix to each reaction tube. | + | Added 49µL Master Mix to each reaction tube.<br> |
| + | |||
---- | ---- | ||
| Line 1,015: | Line 1,035: | ||
(36 cycles) | (36 cycles) | ||
| - | Added 19.5µL Master Mix to each reaction tube. | + | Added 19.5µL Master Mix to each reaction tube.<br> |
| - | Analyzed products on 1% TAE agarose gel which ran for 60 minutes at 100 V. | + | Analyzed products on 1% TAE agarose gel which ran for 60 minutes at 100 V.<br> |
| - | + | <b>Results:</b><br> | |
| + | [[image:Lethbridge_100814PlasmidBackbones.JPG|100px]] | ||
==<font color="white">Aug 14, 2010== | ==<font color="white">Aug 14, 2010== | ||
| Line 1,047: | Line 1,068: | ||
<tr><td>19<td>MT | <tr><td>19<td>MT | ||
<tr><td>20<td>MT | <tr><td>20<td>MT | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</table> | </table> | ||
| Line 1,087: | Line 1,101: | ||
</table> | </table> | ||
| - | + | [[image:Lethbridge_100815MiniprepPCR.JPG|200px]] | |
---- | ---- | ||
| Line 1,119: | Line 1,133: | ||
Incubated at 37<sup>o</sup>C. | Incubated at 37<sup>o</sup>C. | ||
| - | Analyzed results on 2.5% TAE agarose gel which ran at 100 V for 50 minutes. | + | Analyzed results on 2.5% TAE agarose gel which ran at 100 V for 50 minutes.<br> |
| - | + | [[image:Lethbridge_100814ColonyPCRRetryandStartofassembly.JPG|200px]] | |
<b>Results:</b> Lost all the DNA in the column clean-up step and will have to re-do.<br> | <b>Results:</b> Lost all the DNA in the column clean-up step and will have to re-do.<br> | ||
| Line 1,130: | Line 1,144: | ||
<b>Objective:</b> Assemble mms6-dT and lumazine-dT using three antibiotic assembly. <br> | <b>Objective:</b> Assemble mms6-dT and lumazine-dT using three antibiotic assembly. <br> | ||
| - | <b>Method:</b> | + | <b>Method:</b> |
| + | #PCR amplify BioBricks (Prefix/Suffix) | ||
| + | #Restrict BioBricks | ||
| + | #Ligate BioBricks into psB1C3 | ||
| + | #Confirm ligation by PCR analysis (VF2/VR) | ||
| + | #Transform ligation mixes | ||
| + | #Screen colonies with Colony PCR | ||
<u>PCR:</u> Thermocycler set to iGEM program 11<br> | <u>PCR:</u> Thermocycler set to iGEM program 11<br> | ||
| Line 1,170: | Line 1,190: | ||
</table> | </table> | ||
| - | For | + | For pSB1C3 - |
<table><table border ="3"> | <table><table border ="3"> | ||
<tr><td><b>Ingredient</b><td><b>Reaction Mix(µL)</b> | <tr><td><b>Ingredient</b><td><b>Reaction Mix(µL)</b> | ||
| Line 1,258: | Line 1,278: | ||
</table><br> | </table><br> | ||
| - | Added 15 (µL) MM to each tube. | + | Added 15 (µL) MM to each tube.<br> |
| + | <b>Results:</b><br> | ||
==<font color="white">Aug 15, 2010== | ==<font color="white">Aug 15, 2010== | ||
| Line 1,363: | Line 1,384: | ||
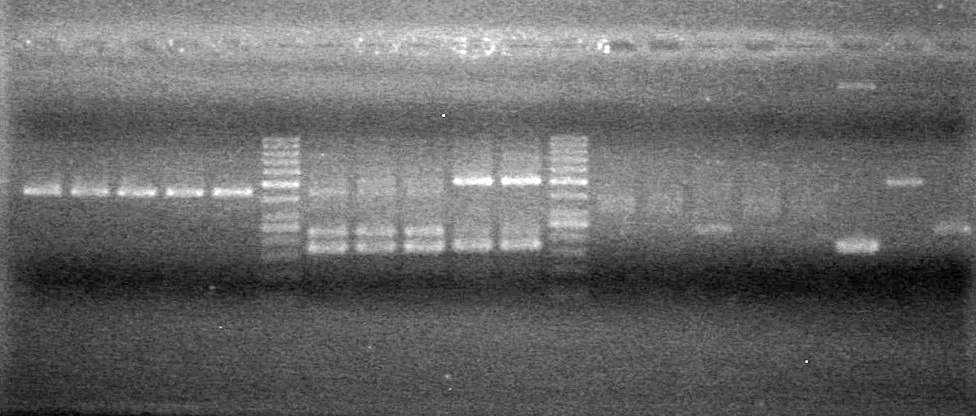
Added 45 (µL) MM to each tube. | Added 45 (µL) MM to each tube. | ||
| - | Analyzed PCR products of BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. | + | Analyzed PCR products of BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. <br> |
| + | <b>Results:</b><br> | ||
| + | [[image:Lethbridge_100815PCRAssembly.JPG|200px]] | ||
| + | |||
| - | |||
---- | ---- | ||
(In Lab: ADS)<br> | (In Lab: ADS)<br> | ||
| Line 1,430: | Line 1,453: | ||
</table><br> | </table><br> | ||
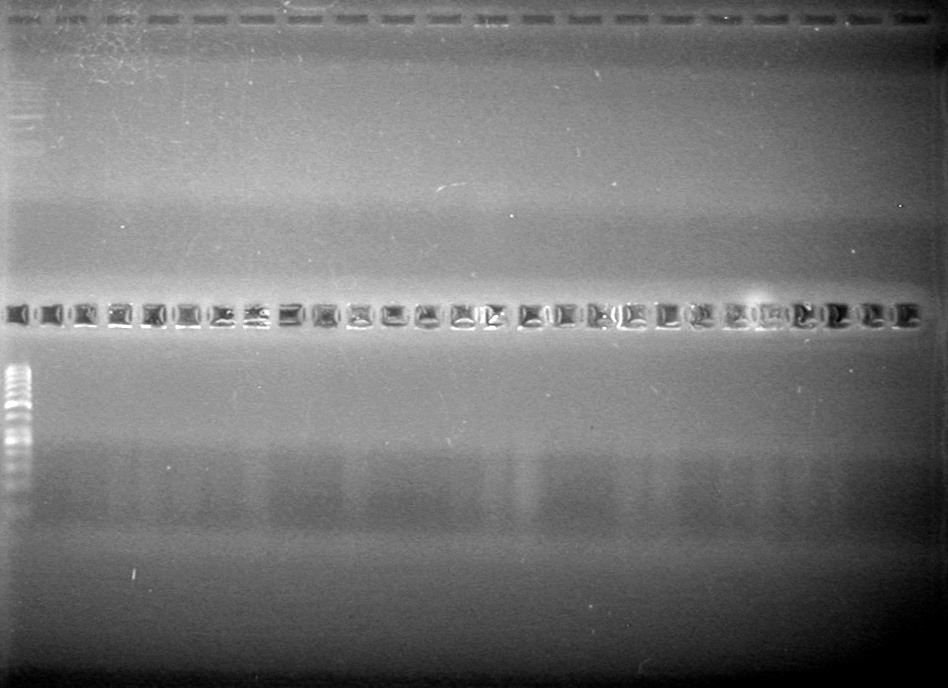
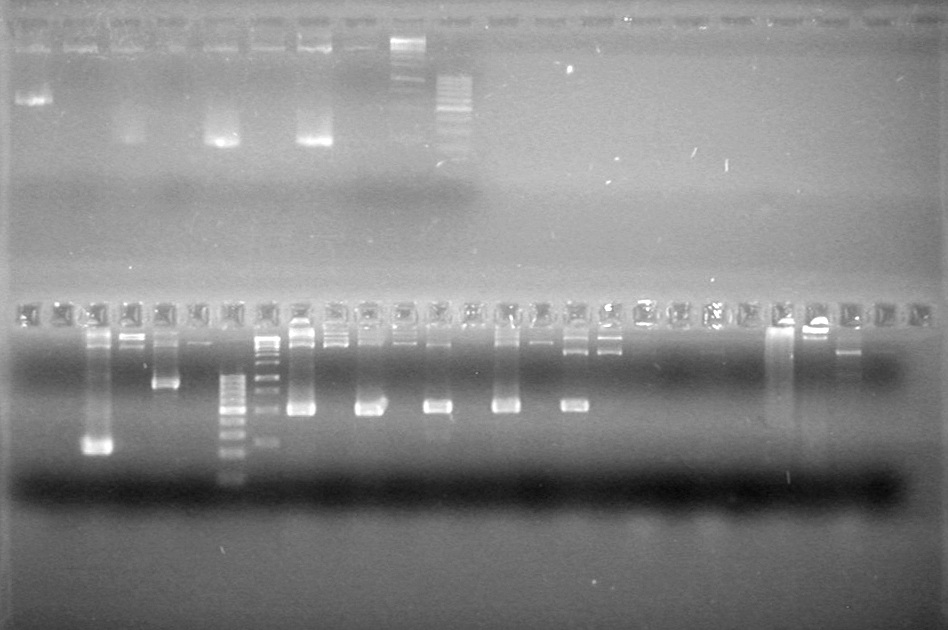
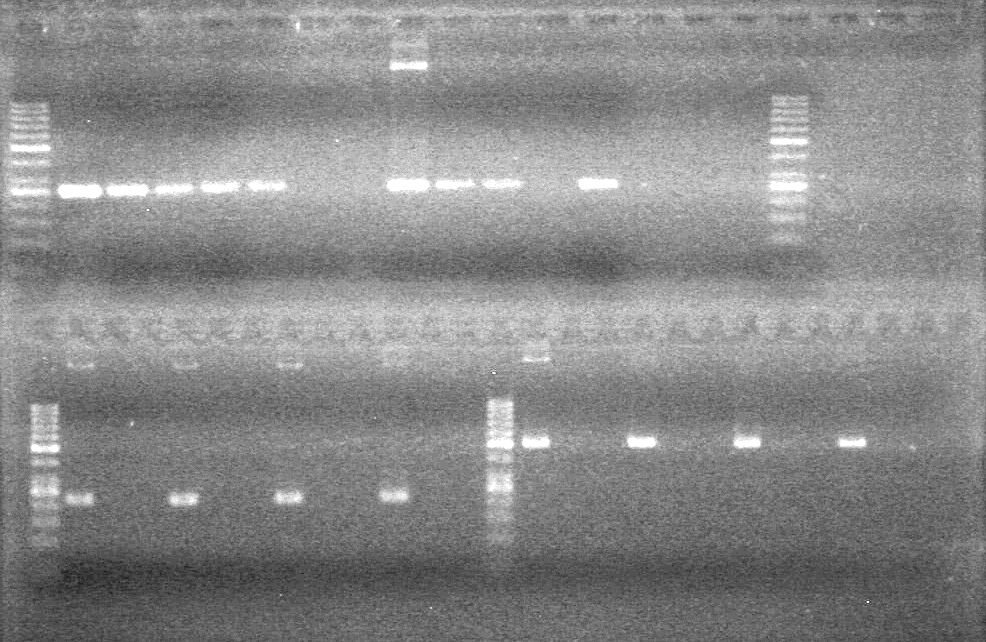
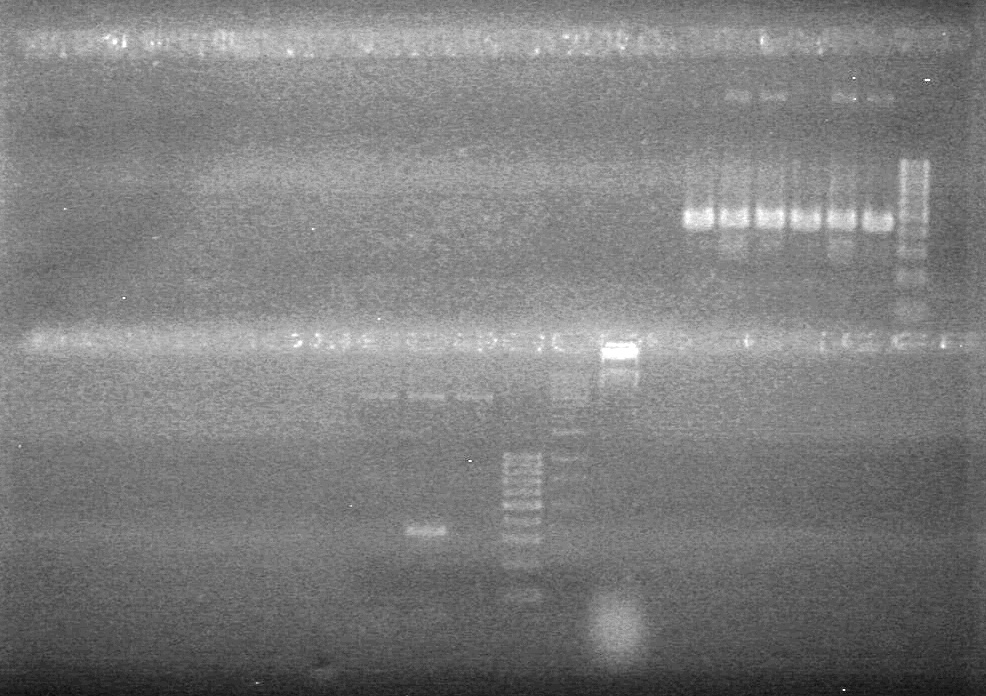
| - | Analyzed PCR products of overnight BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. | + | Analyzed PCR products of overnight BioBrick standard assembly; 3 part (or 3 antibiotic) assembly; and 3 part (3AB) Intermediate/2 part assembly on a 2% TAE agarose gel. <br> |
| - | + | <b>Results:</b><br> | |
| - | + | [[image:Lethbridge_100816PostLigationPCRScreen.JPG|200px]] | |
==<font color="white">Aug 16, 2010 Evening == | ==<font color="white">Aug 16, 2010 Evening == | ||
| Line 1,508: | Line 1,531: | ||
</table><br> | </table><br> | ||
| - | Ran a 2% Agarose gel in 1X TAE buffer for 65 minutes at 100V. | + | Ran a 2% Agarose gel in 1X TAE buffer for 65 minutes at 100V.<br> |
| - | + | [[image:Lethbridge_100817PostLigationPCRTest.JPG|100px]] | |
| - | + | ||
==<font color="white">Aug 17, 2010 Evening == | ==<font color="white">Aug 17, 2010 Evening == | ||
| Line 1,635: | Line 1,657: | ||
Ran on cycle 4 of thermocycler. | Ran on cycle 4 of thermocycler. | ||
| - | Viewed PCRs on 2% TAE agarose gel that ran at 110 V for 30 minutes. | + | Viewed PCRs on 2% TAE agarose gel that ran at 110 V for 30 minutes.<br> |
| - | + | [[image:Lethbridge_100818AssemblyandHeatKillPCR.JPG|200px]] | |
| - | + | ||
---- | ---- | ||
(In Lab: HB)<br> | (In Lab: HB)<br> | ||
| Line 1,729: | Line 1,750: | ||
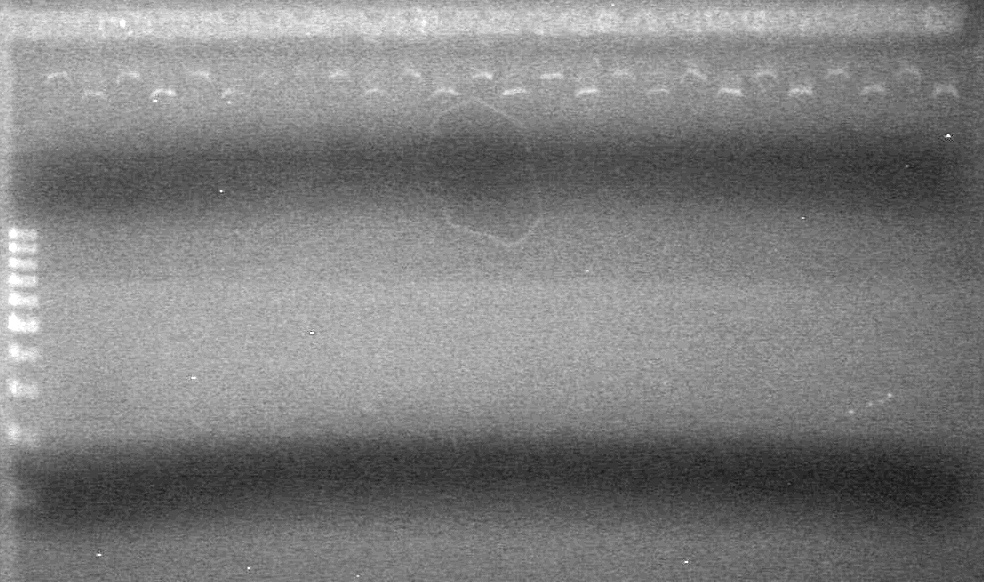
Samples were run on a 2% agarose gel in 1X TAE Buffer. | Samples were run on a 2% agarose gel in 1X TAE Buffer. | ||
| - | |||
| - | |||
---- | ---- | ||
| Line 1,814: | Line 1,833: | ||
---- | ---- | ||
(In Lab: JV)<br> | (In Lab: JV)<br> | ||
| - | Analyzed HB's PCR products on 2% TAE gel which ran for 60 minutes at 100 V. | + | Analyzed HB's PCR products on 2% TAE gel which ran for 60 minutes at 100 V.<br> |
| - | + | [[image:Lethbridge_100819PCRMania.JPG|200px]] | |
| - | + | ||
==<font color="white">Aug 20, 2010== | ==<font color="white">Aug 20, 2010== | ||
| Line 1,828: | Line 1,846: | ||
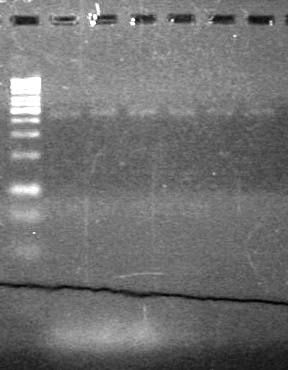
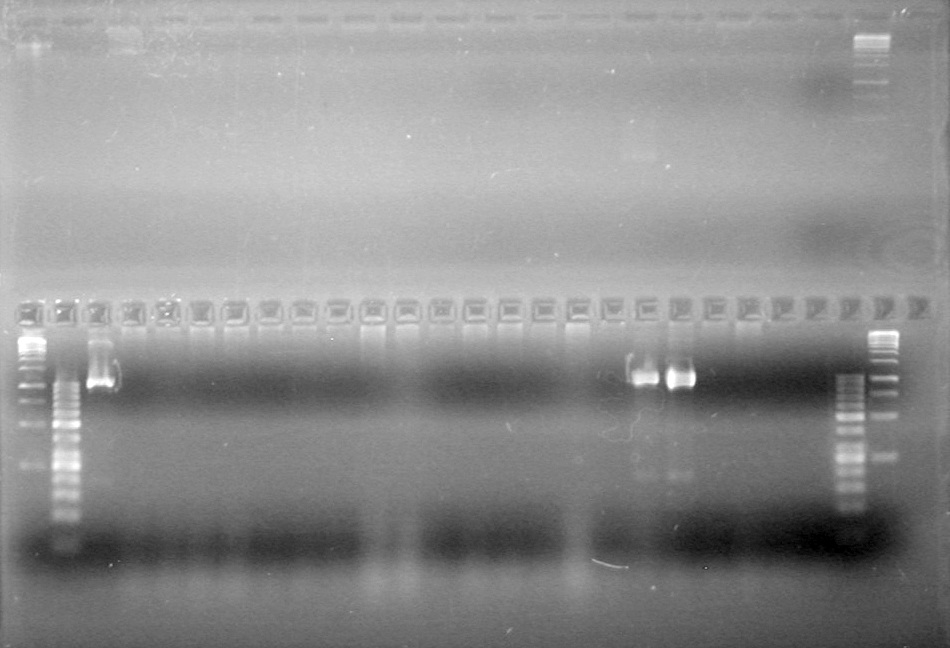
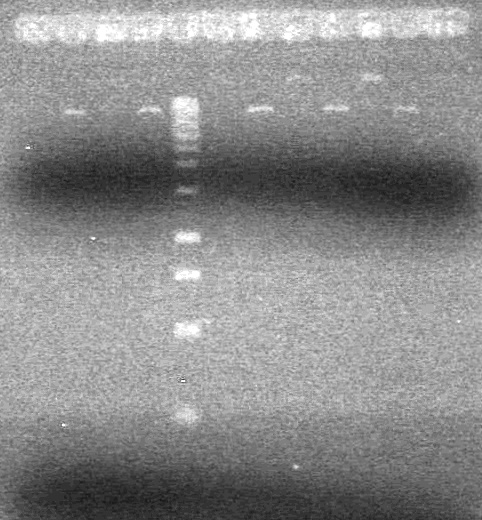
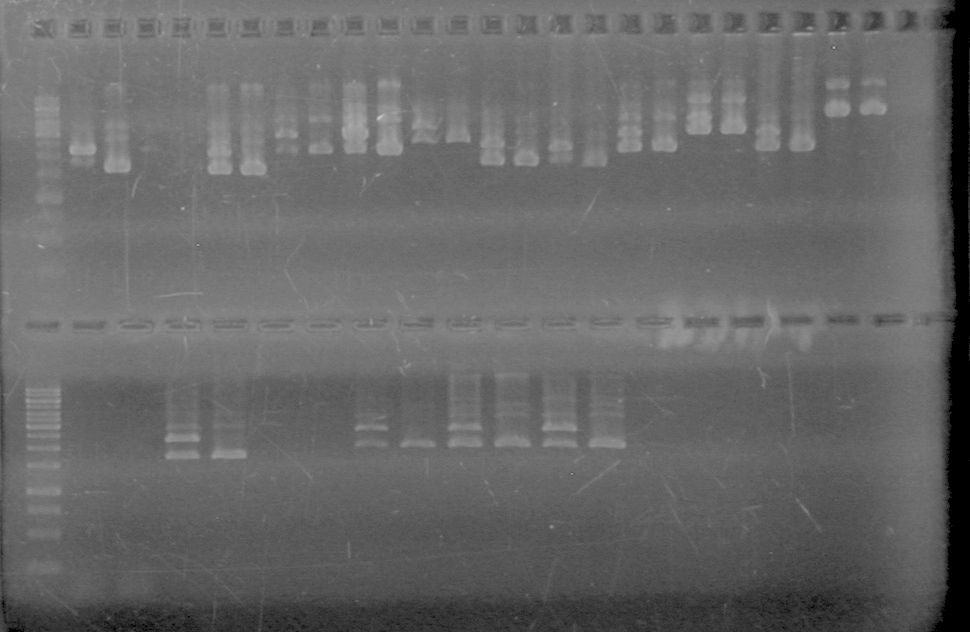
<b>Results:</b> DNA concentration was good, however there was no evidence of an insert into pET-28(A). <br> | <b>Results:</b> DNA concentration was good, however there was no evidence of an insert into pET-28(A). <br> | ||
| - | + | [[image:Lethbridge_100819pET28aTransformScreenGel1.JPG|200px]]<br><br> | |
| + | [[image:Lethbridge_100819pET28aTransformScreenGel2.JPG|200px]]<br><br> | ||
| + | [[image:Lethbridge_100819pET28aTransformScreenGel3.JPG|200px]]<br><br> | ||
| + | [[image:Lethbridge_100819pET28aTransformScreenGel4.JPG|200px]]<br><br> | ||
==<font color="white">Aug 23, 2010== | ==<font color="white">Aug 23, 2010== | ||
| Line 1,864: | Line 1,885: | ||
</table> | </table> | ||
| - | 15(µL) added to each rxn tube. | + | 15(µL) added to each rxn tube.<br> |
| - | Incubated at 37<sup>o</sup>C for 1.5 hours. | + | Incubated at 37<sup>o</sup>C for 1.5 hours. <br> |
| - | + | [[image:Lethbridge_100823HBMaxipreps.jpg|200px]] | |
---- | ---- | ||
(In Lab: AV)<br> | (In Lab: AV)<br> | ||
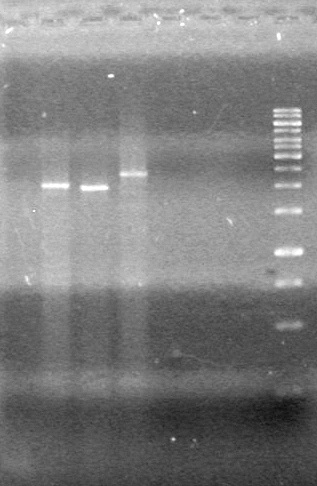
| - | <b>Objective:</b> To PCR amplify | + | <b>Objective:</b> To PCR amplify pSB1T3, pSB1C3, pSB1A3 and run on 1% agarose gel. <br> |
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 1,888: | Line 1,909: | ||
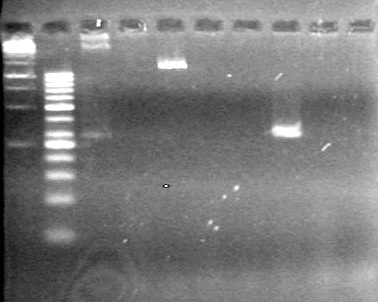
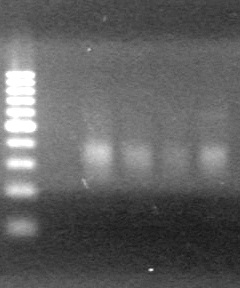
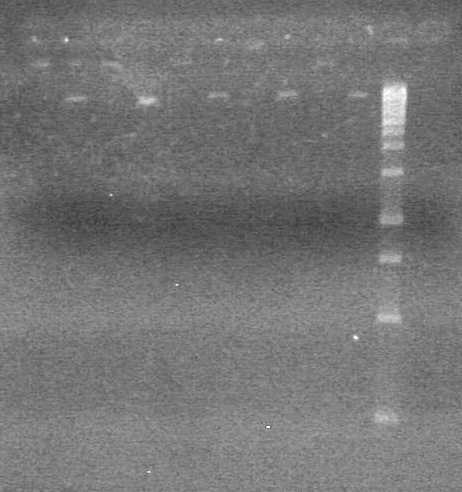
<b>Results:</b> Nothing visible on gel, therefore will have to try loading a larger volume of PCR product.<br> | <b>Results:</b> Nothing visible on gel, therefore will have to try loading a larger volume of PCR product.<br> | ||
| + | [[image:Lethbridge_100823AVBackbonePCR.jpg|100px]] | ||
==<font color="white">Aug 24, 2010== | ==<font color="white">Aug 24, 2010== | ||
| Line 1,945: | Line 1,967: | ||
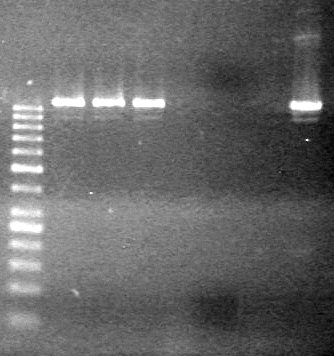
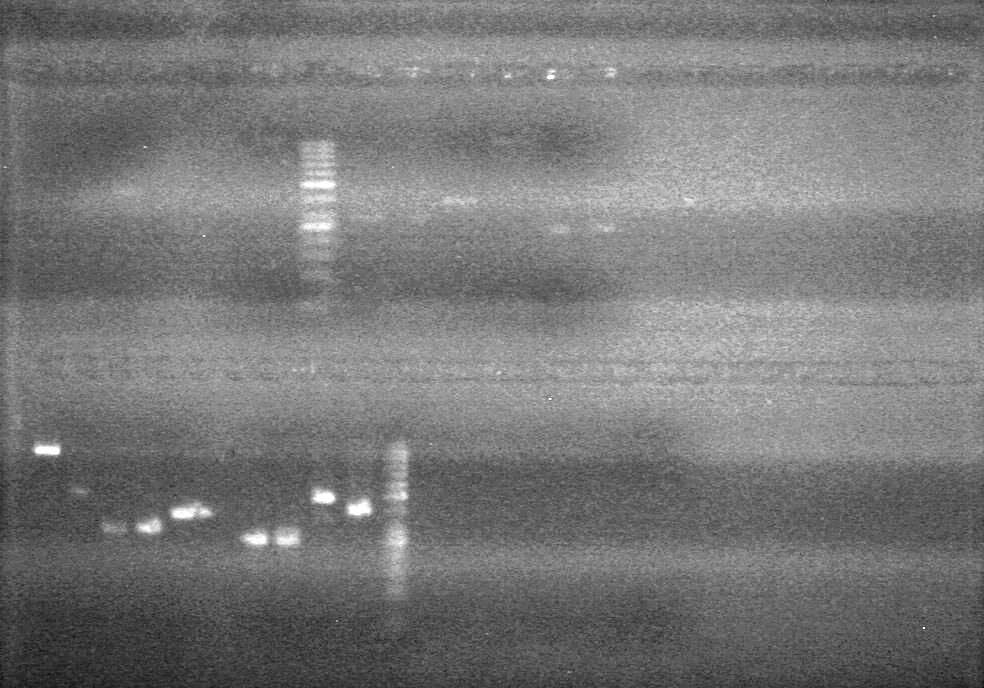
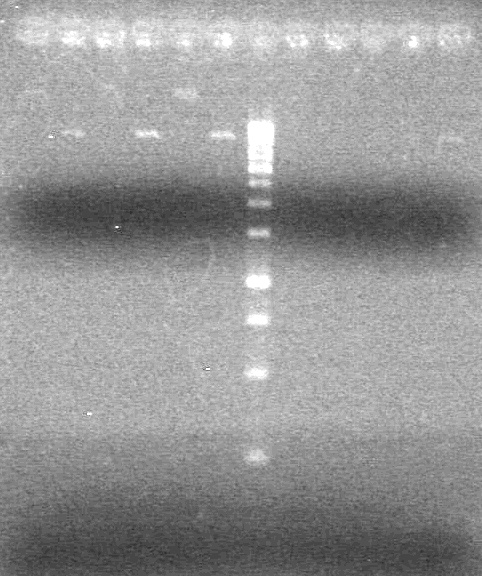
<b>Objective:</b> Run a 2% agarose gel of gradient PCR of xylE. <br> | <b>Objective:</b> Run a 2% agarose gel of gradient PCR of xylE. <br> | ||
| - | + | [[image:Lethbridge_100826 xylE PCR Fix.jpg|200px]] | |
==<font color="white">Aug 26, 2010== | ==<font color="white">Aug 26, 2010== | ||
| Line 1,978: | Line 2,000: | ||
(In Lab: DM)<br> | (In Lab: DM)<br> | ||
<b>Objective:</b> Overexpression test of xylE part (BBa_K118021) in DH5alpha cells in m9 minimal media. <br> | <b>Objective:</b> Overexpression test of xylE part (BBa_K118021) in DH5alpha cells in m9 minimal media. <br> | ||
| + | <b>Method:</b> [[Team:Lethbridge/Notebook/Protocols|Overexpression]] | ||
| + | |||
| + | 0.5 M Catechol was made, to allow for the addition of 1(µL) of this stock solution to be added to the samples taken during the overexpression. | ||
| + | Upon addition of catechol, the solutions turned yellow! | ||
| + | 1 mL samples were taken for SDS-PAGE analysis. | ||
| + | Optical density readings at 600 nm and absorbance readings at 375 and 275 nm were recorded for flasks containing either glucose or sucrose. | ||
| + | ---- | ||
| + | (In Lab: ADS)<br> | ||
| + | <b>Objective:</b> PCR amplify plasmid backbones (pSB1C3, pSB1A3 and pSB1T3). <br> | ||
| + | <b>Method:</b> Use PCR product from Aug 13, 2010 as template DNA. | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Component</b><td><b>1X(µL)</b><td><b>Master Mix(x3.5)(µL)</b> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O<td>14.4<td>52.4 | ||
| + | <tr><td>10x Pfu Buffer with MgSO<sub>4</sub><td>2<td>7 | ||
| + | <tr><td>dNTPs<td>1<td>3.5 | ||
| + | <tr><td>SB-prep-26 Primer<td>0.7<td>2.45 | ||
| + | <tr><td>SB-prep-3P Primer<td>0.7<td>2.45 | ||
| + | <tr><td>Template DNA<td>1<td> | ||
| + | <tr><td>Pfu polymerase<td>0.2<td>0.7 | ||
| + | </table><br> | ||
| + | |||
| + | Added 19(µ:L) to each tube. | ||
| + | Phusion polymerase was used and apparently in the other previously unsuccessful PCRs, instead of Pfu polymerase. | ||
| + | |||
| + | PCR- Conditions: | ||
| + | 1. 95<sup>o</sup>C for 5 min | ||
| + | 2. 94<sup>o</sup>C for 30 sec | ||
| + | 3. 55<sup>o</sup>C for 30 sec | ||
| + | 4. 68<sup>o</sup>C for 4 min | ||
| + | 5. 68<sup>o</sup>C for 10 min | ||
| + | 6. 4<sup>o</sup>C infinitely | ||
| + | (36 cycles) | ||
| + | |||
| + | Analyzed PCR on 1% TAE agarose gel which ran for 60 min at 100 V. | ||
| + | ---- | ||
| + | <b>Objective:</b> Obtain new sources of pSB1T3, pSB1C3 and pSB1A3 plasmid backbone. Backbone from registry used up. <br> | ||
| + | |||
| + | <b>Method:</b> pSB1A3 can be obtained from anything team already possesses. pSB1C3 can be obtained from BBa_J04450 (RFP) in kit. Don't have tetracycline plates so cannot obtain pSB1T3. Used competent cell transformation protocol. <br> | ||
| + | |||
| + | <b>Method:</b> pSB1A3 can be obtained from anything team already possesses. pSB1C3 can be obtained from BBa_J04450 (RFP) in kit. Don't have tetracycline plates so cannot obtain pSB1T3. Used [[Team:Lethbridge/Notebook/Protocols|Competent Cell Transformation]] protocol. <br> | ||
| + | |||
| + | * changes: | ||
| + | **used 50µL aliquottes of DH5&alpha | ||
| + | **did not pipette up and down once, the cells were just swirled 3 times | ||
| + | **added 500µL SOC media, shoock at 37<sup>0</sup>C for 90 min | ||
| + | **platted 250µL and 150µL<br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <td><b>Results</b> | ||
| + | <tr><td>contents<td><b>&250µL</b><td><b>150µL</b> | ||
| + | <tr><td>+ control(pUC19)<td>good<td>good | ||
| + | <tr><td>J04450<td>X (TMTC)<td>X(TMTC) | ||
| + | </table><br> | ||
| + | |||
| + | Prepared overnight cultures for minipreps. Added 5(µL) of 35 mg/mL Chloramphenicl to 5 mL of LB Media. Inoculated media with cells from single colony picked from transformation plate. Incubated overnight at 37<sup>o</sup>C with shaking. | ||
| + | <br><br> | ||
 "
"