Team:UNIPV-Pavia/Calendar/September/settimana5
From 2010.igem.org
m (→October, 1st) |
(→October, 1st) |
||
| (24 intermediate revisions not shown) | |||
| Line 36: | Line 36: | ||
==September, 27th== | ==September, 27th== | ||
| + | <font size=4>[[Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/test27settembre|Tecan Test]]</font> | ||
| + | |||
| + | ---- | ||
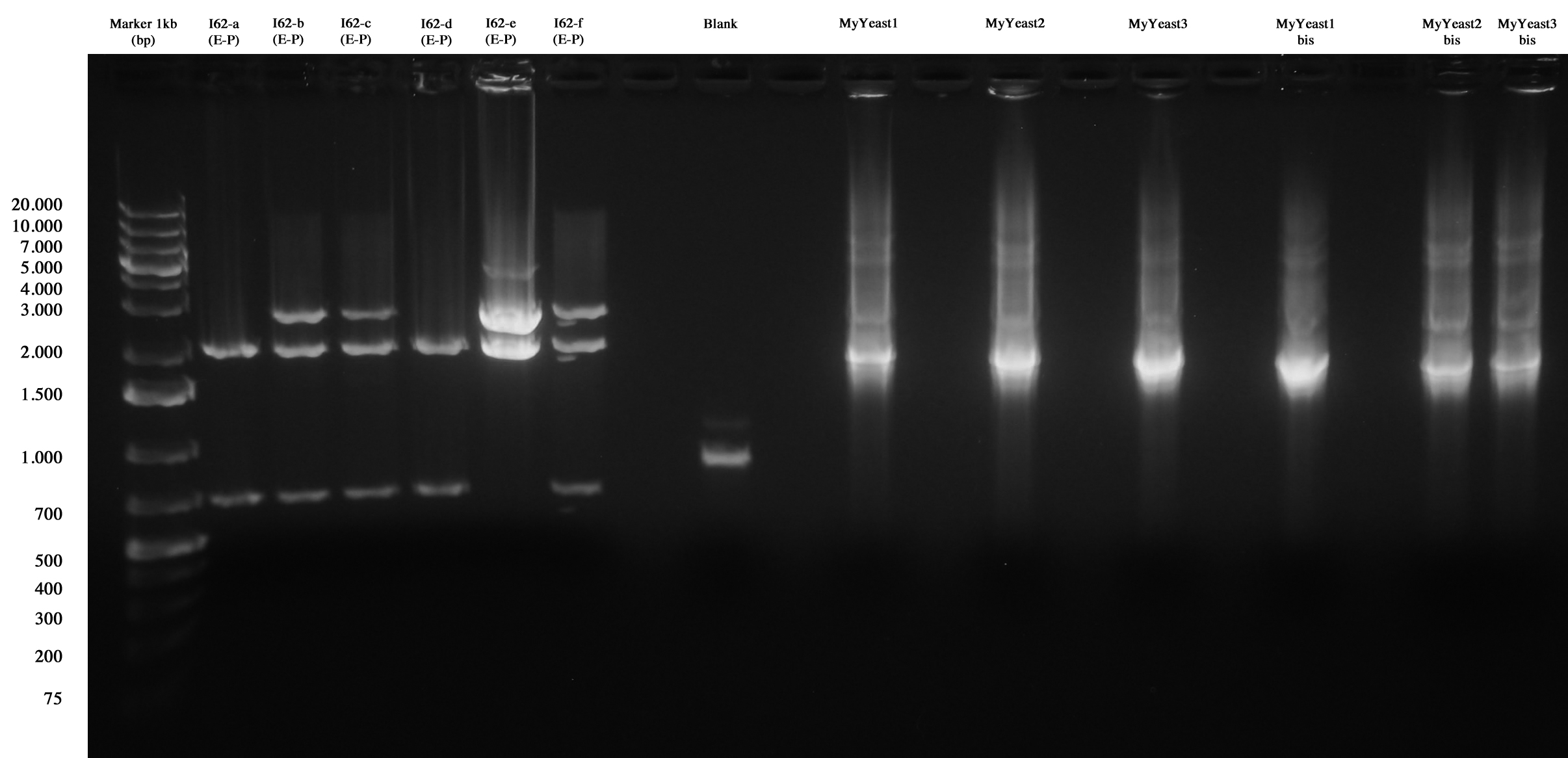
MiniPrep and E-P digest for I62-a, I62-b, I62-c, I62-d, I62-e, I62-f | MiniPrep and E-P digest for I62-a, I62-b, I62-c, I62-d, I62-e, I62-f | ||
| - | DNA | + | DNA PCR for MyYeast 1-2-3 |
Gel run | Gel run | ||
| Line 48: | Line 51: | ||
| - | Protein electrophoresis for phasins | + | Protein electrophoresis for phasins. |
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==September, 28th== | ==September, 28th== | ||
| - | <font size=4>[[Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/test28settembre|Tecan Test]]</font> | + | <font size=4>[[Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/test28settembre|Tecan Test]]</font> |
---- | ---- | ||
| Line 76: | Line 79: | ||
==September, 29th== | ==September, 29th== | ||
| - | |||
| - | |||
Colony PCR for 6 newly picked colonies. | Colony PCR for 6 newly picked colonies. | ||
[[Image:UNIPV10_29_09_2010_I55_screen.jpg|thumb|200px|center|I55 colony PCR screening.]] | [[Image:UNIPV10_29_09_2010_I55_screen.jpg|thumb|200px|center|I55 colony PCR screening.]] | ||
| Line 108: | Line 109: | ||
|} | |} | ||
| + | ---- | ||
| + | Inoculum of MC43, MG43 in 5ml LB with chloramphenicol concentration of 12.5 ug/ml. | ||
| Line 114: | Line 117: | ||
==September, 30th== | ==September, 30th== | ||
| - | <font size=4>[[Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/ | + | <font size=4>[[Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/test30settembre|Tecan Test]]</font> |
---- | ---- | ||
| Line 162: | Line 165: | ||
Cultures were grown ON at 37°C 220 rpm. | Cultures were grown ON at 37°C 220 rpm. | ||
| - | Tomorrow we will test the ability of these | + | Tomorrow we will test the ability of these strains to produce GFP and to lyse when HSL is added. |
| + | |||
| + | ---- | ||
| + | In order to excise the chloramphenicol resistance cassette from the integrants in E.coli genome we transformed the cells with a plasmid (pCP20 plasmid) carrying the gene encoding for the flippase protein. This protein is able to cut fragment between two FRT flanking sequences. pCP20 was transformed in MC43-a and MG43-a and plated on LB+Amp agar plate. | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
==October, 1st== | ==October, 1st== | ||
| + | |||
| + | <font size=4>[[Team:UNIPV-Pavia/Material Methods/Measurements/Tecan/test1ottobre|Tecan Test]]</font> | ||
| + | |||
| + | ---- | ||
This morning, we checked tranformed plates: | This morning, we checked tranformed plates: | ||
| Line 223: | Line 233: | ||
*INTEIN_C3: INTEIN (E-P) + <partinfo>pSB1C3</partinfo> (E-P) | *INTEIN_C3: INTEIN (E-P) + <partinfo>pSB1C3</partinfo> (E-P) | ||
| + | ---- | ||
| + | Single colony was picked from MC43 and MG43 plates and inoculated in 5 ml of LB and incubated at 37°C, 220 rpm ON. | ||
| + | 12 hours late 5 ul were streaked on non selective LB plates and incubated at 43°C ON in order to ensure the loss of pCP20 plasmid. | ||
| Line 247: | Line 260: | ||
1,5 hours E-P digest (for screening) and gel run | 1,5 hours E-P digest (for screening) and gel run | ||
| - | [[Image:UNIPV10_02_10_2010_I73_screening.jpg|thumb|200px|center|I70 | + | [[Image:UNIPV10_02_10_2010_I73_screening.jpg|thumb|200px|center|I70, I71, I73, A9_4C5, A99_4C5 screening.]] |
As you can see they are (except for I73-6) all positive. So we stocked I73-1 and stored it at -80°C. | As you can see they are (except for I73-6) all positive. So we stocked I73-1 and stored it at -80°C. | ||
| Line 256: | Line 269: | ||
---- | ---- | ||
Inoculum of I55-1 into 5 ml LB+Amp. ON growth at +37°C, 220 rpm. | Inoculum of I55-1 into 5 ml LB+Amp. ON growth at +37°C, 220 rpm. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | 3 colonies each plate were picked from MC43_flip and MG43_flip plates and inoculated in 5 ml of non selective LB. To confirm the loss of the chloramphenicol resistance cassette and of the pCP20 plasmid, each colony was inoculated also in 5 ml of LB+Cm12.5 and 5 ml of LB+Amp. | ||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
| Line 276: | Line 293: | ||
---- | ---- | ||
| - | Colony PCR for I74 | + | Colony PCR for I74-1..6. |
---- | ---- | ||
Gel run for PCR samples and I55 (E-S) and cut for this one. | Gel run for PCR samples and I55 (E-S) and cut for this one. | ||
[[Image:UNIPV10_03_10_2010_I74_screen_I55_cut.jpg|thumb|200px|center|I74 screening and I55 E-S cut.]] | [[Image:UNIPV10_03_10_2010_I74_screen_I55_cut.jpg|thumb|200px|center|I74 screening and I55 E-S cut.]] | ||
| - | Very bad PCR. We performed a massive PCR from colony during the night (I74-7 | + | Very bad PCR. We performed a massive PCR from colony during the night (I74-7..24), we ran it the following day. |
Gel extraction and quantification: | Gel extraction and quantification: | ||
| Line 291: | Line 308: | ||
---- | ---- | ||
Inoculum of I35 (last quantification after gel extraction was too poor; we will digest it again E-P) and I54 into 5 ml LB+Amp: ON growth at +37°C, 220 rpm. | Inoculum of I35 (last quantification after gel extraction was too poor; we will digest it again E-P) and I54 into 5 ml LB+Amp: ON growth at +37°C, 220 rpm. | ||
| + | |||
| + | ---- | ||
| + | Each culture of MC43_flip and MG43_flip in selective LB didn't grown, confirming the loss of pCP20 plasmid and of the chloramphenicol resistance cassette integrated into the E.coli genome. | ||
| + | |||
<div align="right"><small>[[#indice|^top]]</small></div> | <div align="right"><small>[[#indice|^top]]</small></div> | ||
Latest revision as of 18:36, 27 October 2010
|
|
||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||
 "
"