Team:Imperial College London/Results/Exp5
From 2010.igem.org
(New page: {{:Team:Imperial_College_London/Templates/Header}} {{:Team:Imperial_College_London/Templates/ResultsHeader}}) |
|||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College_London/Templates/Header}} | {{:Team:Imperial_College_London/Templates/Header}} | ||
{{:Team:Imperial_College_London/Templates/ResultsHeader}} | {{:Team:Imperial_College_London/Templates/ResultsHeader}} | ||
| + | |||
| + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;" colspan="2"|Experiment 5 | Characterizing GFP-XylE gene product in whole cell cultures | ||
| + | |- | ||
| + | |colspan="2"| | ||
| + | As part of the project, the team needed to identify a reporter gene that would give us the visual output. However the means of getting this visual output needed to be inducible, and not constitutively active, when the appropriate input is present. Another specification for this reporter was that it's activation did not to go through the usual route of transcription/translation, which is a slow process. We had little success of finding something suitable for our project so we decided to build our own novel system. We decided to use the gene product of XylE, catechol 2,3 dioxygenase (C2,3O). By looking at three dimensional diagrams of C2,3O and using software tools we proposed an inducible model of this enzyme. | ||
| + | |- | ||
| + | | | ||
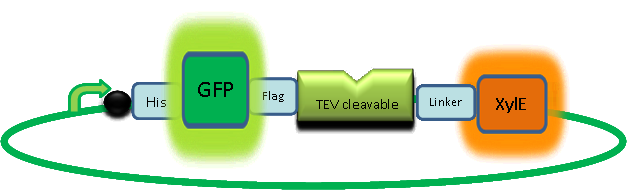
| + | In our construct, GFP was added to the N-terminus of C2,3O monomer. The C2,3O enzyme is an obligate homotetramer where all 4 subunits are required for its activity. We hoped that by adding GFP at the N-terminus of the each monomer, we would block the tetramerization of the enzyme, thus rendering it inactive. This modification is important as it makes our system inducible. The cleavage of the GFP by TEV protease, which is synthesised on arrival of the input signal, means that Catechol dioxygenase becomes active only when the parasite is detected. | ||
| + | | | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:399px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:IC Module3.JPG|395px]] | ||
| + | |- | ||
| + | |The GFP-XylE gene construct. | ||
| + | |} | ||
| + | </div> | ||
| + | |- | ||
| + | |colspan="2"| | ||
| + | |||
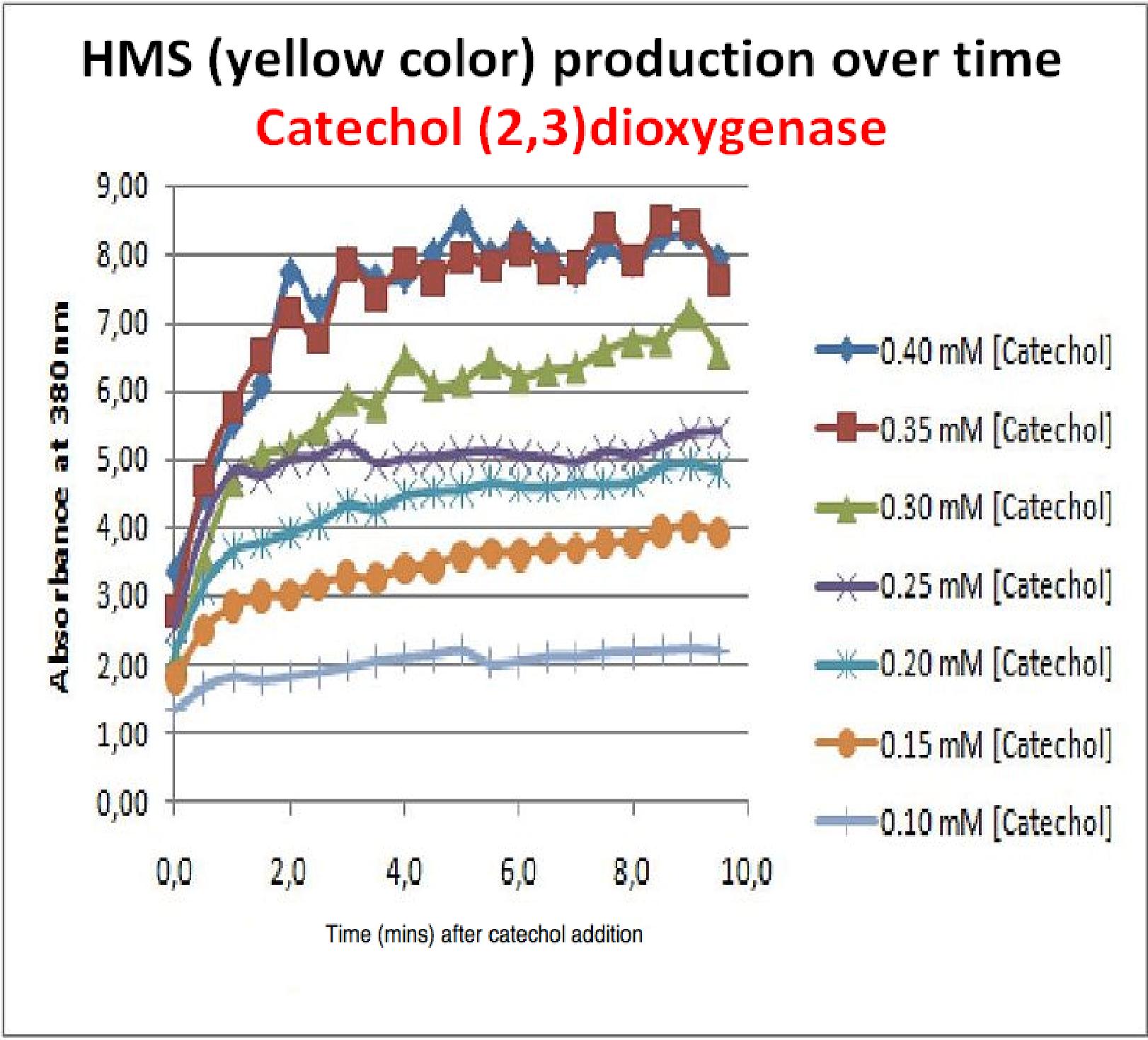
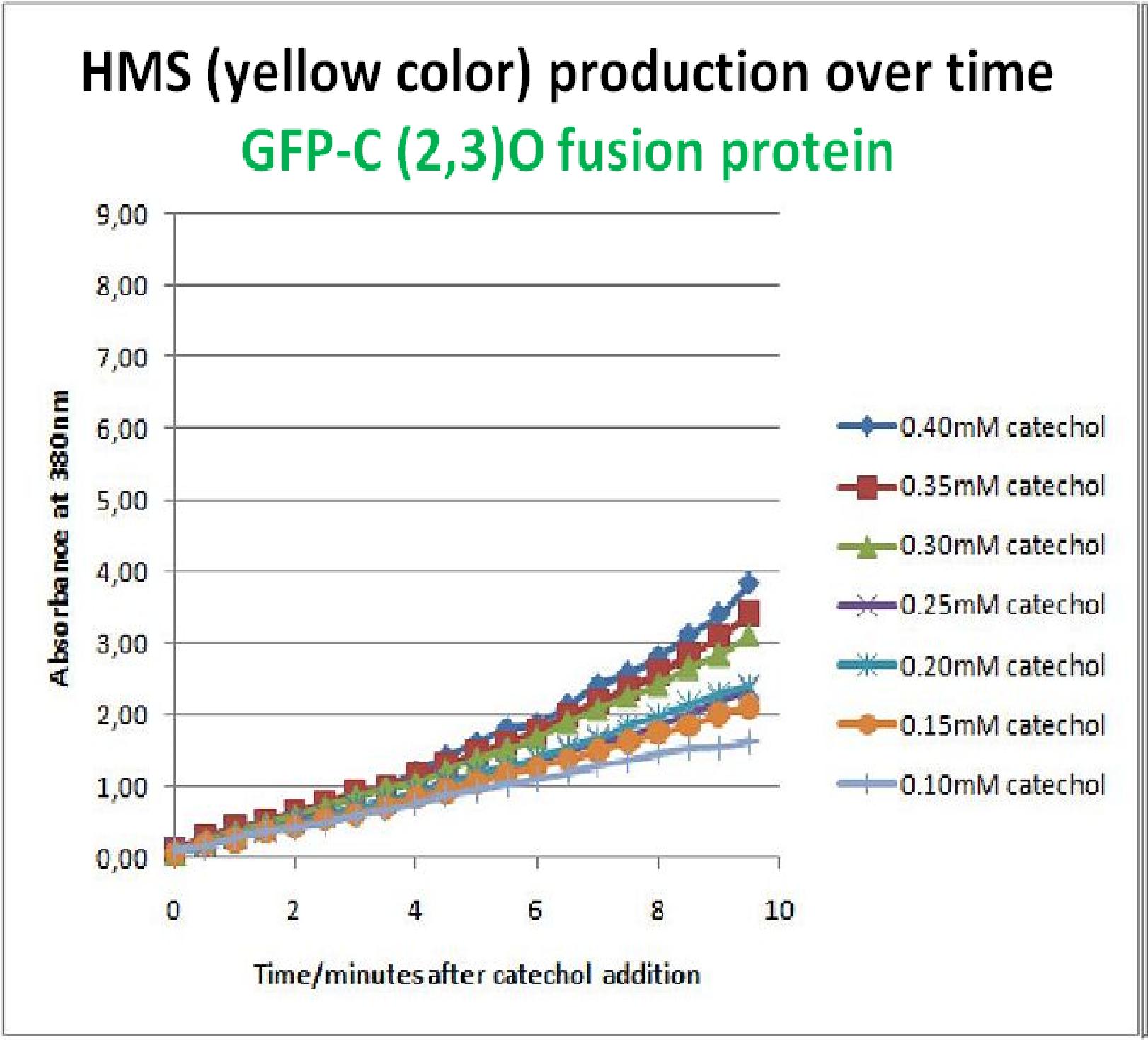
| + | To investigate this, an experiment was designed where catechol was added to GFP-XylE transformed ''E.coli''. The progress of the reaction was monitored by measuring production of the 2-hydroxymuconic semialdehyde (yellow), which arbsorbs light at 380nm. The product concentraton correlates well to the velocity of the reaction, if divided by time, and by using various initial catechol concentrations as a substrate, a kinetic profile of the reaction could be constructed. The experimental set up is essentially the same as described above for determining the kinetics of C2,3O. The only difference is that the ''E.coli'' cells used were the ones transformed with GFP-XylE gene construct instead of XylE gene. | ||
| + | |||
| + | The results of the assay show conclusively that the N-terminal fusion of GFP gene to the XylE gene affects activity of the wild type C2,3O. Although the activity is leaky, producing very small amounts of the colored product of the reaction in the presence of catechol, the alteration in the reaction profile of the reaction is impressively large. The two graphs below are equivalent and comparable in the sense that the only different between these experiments is the ''E.coli'' cells used. The left graph represents cells expressing the original XylE gene, while the graph on the right hand side represents cultures expressing the GFP-XylE gene. These genes are both under the influence of the Pveg promoter so that we can assume equal production of protein product. | ||
| + | |- | ||
| + | | | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:399px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:HMS prod. over time curve for xylE.jpg|395px]] | ||
| + | |- | ||
| + | |Graph shows production of HMS (yellow product) over time after catechol addition at time 0 minutes. Different curves represent different catechol concentration added to the cell cultures. | ||
| + | |} | ||
| + | </div> | ||
| + | | | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:399px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Reaction profile of GFP-XylE.jpg|395px]] | ||
| + | |- | ||
| + | |Graph shows production of HMS over time after catechol addition at time 0 minutes. Different colored curves represent different substrate concentrations added at time 0min in cell cultures. | ||
| + | |} | ||
| + | </div> | ||
| + | |- | ||
| + | |colspan="2"| | ||
| + | If we take 3 minutes as the reference point on the graphs, as this is where the XylE expressing cells saturate the system with the colored product, we can see that there is about '''a 10-fold reduction in the rate at which the protein product of GFP-XylE gene construct catalyses the reaction in comparison to the wild type XylE gene'''. From this we can deduce that the attached GFP protein influences tetramerisation of the Catechol dioxygenase enzyme, so that the equilibrium: | ||
| + | 4 monomers (inactive) <-----> tetramer (active) | ||
| + | is shifted is shifted towards protein remaining as monomers and thus inactive. | ||
| + | |||
| + | The results of this experiment were really encouraging for the team, as it shows that we have managed to inactivate the normally constitutively active reporter protein of our output module. The lab team now proposes an experiment where the inactive fusion protein (GFP-catechol dioxygenase) is exposed to TEV protease. Reversion of the reaction profile back to the kinetics of the wild type catechol dioxygenase would mean that we have successfully constructed an inducible reporter system. | ||
| + | |||
| + | |} | ||
Revision as of 15:12, 27 October 2010
| Experimental Results | Exp 1 | Exp 2 | Exp 3 | Exp 4 | Exp 5 | Exp 6 | Exp 7 |
| Testing is a fundamental stage of the engineering design cycle and is a crucial part of charactrising BioBrick Standard Biological Parts so that other people can benefit from our work. We've compiled all our results on this page, detailing how the experiments were carried out and the significance of the data. | |
| Experiment 5 | Characterizing GFP-XylE gene product in whole cell cultures | |
|
As part of the project, the team needed to identify a reporter gene that would give us the visual output. However the means of getting this visual output needed to be inducible, and not constitutively active, when the appropriate input is present. Another specification for this reporter was that it's activation did not to go through the usual route of transcription/translation, which is a slow process. We had little success of finding something suitable for our project so we decided to build our own novel system. We decided to use the gene product of XylE, catechol 2,3 dioxygenase (C2,3O). By looking at three dimensional diagrams of C2,3O and using software tools we proposed an inducible model of this enzyme. | |
|
In our construct, GFP was added to the N-terminus of C2,3O monomer. The C2,3O enzyme is an obligate homotetramer where all 4 subunits are required for its activity. We hoped that by adding GFP at the N-terminus of the each monomer, we would block the tetramerization of the enzyme, thus rendering it inactive. This modification is important as it makes our system inducible. The cleavage of the GFP by TEV protease, which is synthesised on arrival of the input signal, means that Catechol dioxygenase becomes active only when the parasite is detected. | |
|
To investigate this, an experiment was designed where catechol was added to GFP-XylE transformed E.coli. The progress of the reaction was monitored by measuring production of the 2-hydroxymuconic semialdehyde (yellow), which arbsorbs light at 380nm. The product concentraton correlates well to the velocity of the reaction, if divided by time, and by using various initial catechol concentrations as a substrate, a kinetic profile of the reaction could be constructed. The experimental set up is essentially the same as described above for determining the kinetics of C2,3O. The only difference is that the E.coli cells used were the ones transformed with GFP-XylE gene construct instead of XylE gene. The results of the assay show conclusively that the N-terminal fusion of GFP gene to the XylE gene affects activity of the wild type C2,3O. Although the activity is leaky, producing very small amounts of the colored product of the reaction in the presence of catechol, the alteration in the reaction profile of the reaction is impressively large. The two graphs below are equivalent and comparable in the sense that the only different between these experiments is the E.coli cells used. The left graph represents cells expressing the original XylE gene, while the graph on the right hand side represents cultures expressing the GFP-XylE gene. These genes are both under the influence of the Pveg promoter so that we can assume equal production of protein product. | |
|
If we take 3 minutes as the reference point on the graphs, as this is where the XylE expressing cells saturate the system with the colored product, we can see that there is about a 10-fold reduction in the rate at which the protein product of GFP-XylE gene construct catalyses the reaction in comparison to the wild type XylE gene. From this we can deduce that the attached GFP protein influences tetramerisation of the Catechol dioxygenase enzyme, so that the equilibrium: 4 monomers (inactive) <-----> tetramer (active) is shifted is shifted towards protein remaining as monomers and thus inactive. The results of this experiment were really encouraging for the team, as it shows that we have managed to inactivate the normally constitutively active reporter protein of our output module. The lab team now proposes an experiment where the inactive fusion protein (GFP-catechol dioxygenase) is exposed to TEV protease. Reversion of the reaction profile back to the kinetics of the wild type catechol dioxygenase would mean that we have successfully constructed an inducible reporter system. | |
 "
"