Team:Lethbridge/Notebook/Lab Work/June
From 2010.igem.org
JustinVigar (Talk | contribs) (→June 3/2010) |
Adam.smith4 (Talk | contribs) (→June 28/2010) |
||
| (131 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | <div style="background-color:#000000; color:white"> | |
| + | <html> | ||
| - | + | <br> | |
| - | + | <table border="0" width="100%" style="background-color:#000000"> | |
| - | + | <tr> | |
| - | + | <th> | |
| - | + | <image src="https://static.igem.org/mediawiki/2010/2/29/UofLteamlogo.jpg" width="200px"/> | |
| - | + | ||
| - | + | </th> | |
| - | + | ||
| - | + | ||
| - | + | <th> | |
| - | + | ||
| - | + | ||
| - | + | <image src="https://static.igem.org/mediawiki/2010/9/91/UofLLabWork.JPG" height="300px"/> | |
| - | + | ||
| - | + | ||
| - | + | </th> | |
| - | + | ||
| - | + | ||
| - | + | <th> | |
| - | + | ||
| - | + | ||
| - | + | <image src="https://static.igem.org/mediawiki/2010/2/29/UofLteamlogo.jpg" width="200px"/> | |
| - | + | ||
| - | + | ||
| - | + | </th> | |
| - | + | </tr> | |
| - | == | + | |
| - | ===June 1/2010=== | + | </table> |
| + | |||
| + | |||
| + | |||
| + | <br> | ||
| + | |||
| + | <align="centre"> | ||
| + | <table border="0" width="100%" style="background-color:#000000"> | ||
| + | |||
| + | <tr> | ||
| + | |||
| + | <th> | ||
| + | |||
| + | <a href="https://2010.igem.org/Team:Lethbridge"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/2/22/UofLHome.jpg" width="80"/> | ||
| + | </a> | ||
| + | |||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Team"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/0d/UofLTeam.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Project"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/8d/UofLProjectbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLNotebookbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Parts"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/84/UofLPartsSubmittedToTheRegistrybutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Modeling"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/e/e1/UofLModelingbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Ethics"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/2/26/UofLEthicsbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Safety"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/00/UofLSafetybutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Art"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/0/0a/UofLArt.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/News"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/c/c3/UofLNewsButton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | </table> | ||
| + | </body> | ||
| + | </html> | ||
| + | <hr> | ||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <font color="white">Feel free to look around our notebook! | ||
| + | </center> | ||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
| + | <center> | ||
| + | <table border="0" width="28%" style="background-color:#000000"> | ||
| + | |||
| + | <tr> | ||
| + | <th> | ||
| + | <div class="miniBar"> | ||
| + | <div class="countdown"><object type="application/x-shockwave-flash" data="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=10&date_day=27&date_year=0&un=THE WIKI FREEZE&size=normal&mo=10&da=27&yr=2010" width="300" height="100"><param name="movie" value="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=10&date_day=27&date_year=0&un=THE WIKI FREEZE&size=normal&mo=10&da=27&yr=2010" /><param name="bgcolor" value="#000000" /></object><img src="http://www.oneplusyou.com/q/img/bb_badges/countdown.jpg" alt="" style="display: none;" height="1" width="1" /></div> | ||
| + | <div class="miniContainer"> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLNotebookbutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Protocols"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/9/91/UofLprotocolsbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Calendar"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/7/73/UofLcalendar.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th> | ||
| + | <div class="miniBar"> | ||
| + | <div class="countdown"><object type="application/x-shockwave-flash" data="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=11&date_day=05&date_year=0&un=THE IGEM JAMBOREE&size=normal&mo=11&da=05&yr=2010" width="300" height="100"><param name="movie" value="http://www.oneplusyou.com/bb/files/countdown/countdown.swf?co=FFFFFF&bgcolor=000000&date_month=11&date_day=05&date_year=0&un=THE IGEM JAMBOREE&size=normal&mo=11&da=05&yr=2010" /><param name="bgcolor" value="#000000" /></object><img src="http://www.oneplusyou.com/q/img/bb_badges/countdown.jpg" alt="" style="display: none;" height="1" width="1" /></div> | ||
| + | <div class="miniContainer"> | ||
| + | </th> | ||
| + | <tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | <hr> | ||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <font color="white">Here you can check out the work we have done in the lab, click on a month to take a look! | ||
| + | </center> | ||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | <body> | ||
| + | <center> | ||
| + | <table border="0" width="50%" style="background-color:#000000"> | ||
| + | |||
| + | <tr> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/April"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/8a/UofLapril.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/May"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/7/7b/UofLmaybutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/June"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/8/80/UofLjunebutton.jpg" width="80"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/July"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/5/53/UofLjulybutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/August"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/1/15/UofLaugustbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/September"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/4/4d/UofLseptemberbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | <th><a href="https://2010.igem.org/Team:Lethbridge/Notebook/Lab_Work/October"> | ||
| + | <img src="https://static.igem.org/mediawiki/2010/4/4e/UofLoctoberbutton.jpg" width="60"/> | ||
| + | </a> | ||
| + | </th> | ||
| + | |||
| + | |||
| + | |||
| + | <tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | </body> | ||
| + | </html> | ||
| + | <hr> | ||
| + | |||
| + | <BLOCKQUOTE> | ||
| + | |||
| + | =<font color="white">June 2010= | ||
| + | ==<font color="white">June 1/2010== | ||
JV quantified the amount of DNA in gels run to date using ImageJ software. Results to be posted in [[Team:Lethbridge/Notebook/Working_Plasmids|working plasmids box]].<br> | JV quantified the amount of DNA in gels run to date using ImageJ software. Results to be posted in [[Team:Lethbridge/Notebook/Working_Plasmids|working plasmids box]].<br> | ||
| Line 57: | Line 218: | ||
*BglII Endonuclease (Bba_K112106)<br> | *BglII Endonuclease (Bba_K112106)<br> | ||
| - | ===June 2/2010 | + | ==<font color="white">June 2/2010== |
(In Lab: JV)<br> | (In Lab: JV)<br> | ||
| Line 130: | Line 291: | ||
Killed enzymes by incubating reactions for 10 minutes at 80<sup>o</sup>C</ul> | Killed enzymes by incubating reactions for 10 minutes at 80<sup>o</sup>C</ul> | ||
| - | ===June 2/2010 - Evening | + | ==<font color="white">June 2/2010 - Evening== |
<b>Objective:</b> Set up new ligations of pLacI and sRBS-Lum-dT according to Tom Knight's protocol. Previous ligation had very little DNA.<br> | <b>Objective:</b> Set up new ligations of pLacI and sRBS-Lum-dT according to Tom Knight's protocol. Previous ligation had very little DNA.<br> | ||
<b>Relevant Information:</b><br> | <b>Relevant Information:</b><br> | ||
| Line 179: | Line 340: | ||
Incubate for 20 minutes at 80<sup>o</sup>C to heat kill<br> | Incubate for 20 minutes at 80<sup>o</sup>C to heat kill<br> | ||
| - | ===June 3/2010 | + | ==<font color="white">June 3/2010== |
Carried out protocol described in June 2/2010 - Evening<br> | Carried out protocol described in June 2/2010 - Evening<br> | ||
Analyzed results on 1% agarose gel.Load order as follows:<br> | Analyzed results on 1% agarose gel.Load order as follows:<br> | ||
| Line 209: | Line 370: | ||
| - | ===June 3/2010 - Evening | + | ==<font color="white">June 3/2010 - Evening== |
<b>Objective:</b> Repeat restriction of pSB1T3 and ligate with pLacI and sRBS-Lum-dT. Previous ligations all used up on gel.<br> | <b>Objective:</b> Repeat restriction of pSB1T3 and ligate with pLacI and sRBS-Lum-dT. Previous ligations all used up on gel.<br> | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 222: | Line 383: | ||
Incubate for 30 minutes at room temperature to ligate<br> | Incubate for 30 minutes at room temperature to ligate<br> | ||
Incubate for 20 minutes at 80<sup>o</sup>C to heat kill<br> | Incubate for 20 minutes at 80<sup>o</sup>C to heat kill<br> | ||
| - | Following ligation, transformed using | + | Following ligation, transformed using [[Team:Lethbridge/Notebook/Protocols|transformation protocol]]. |
Plates incubated in 37<sup>o</sup>C incubator for 44 hours. <br> | Plates incubated in 37<sup>o</sup>C incubator for 44 hours. <br> | ||
| - | <b>Results:</b> Only plate pLacI (D6) + sRBS-Lum-dT (G2) + pSB1T3 grew; had 2 colonies. Control plate did not grow, acidentally plated on tetracycline plate instead of ampicillin (pUC19). | + | <b>Results:</b> Only plate pLacI (D6) + sRBS-Lum-dT (G2) + pSB1T3 grew; had 2 colonies. Control plate did not grow, acidentally plated on tetracycline plate instead of ampicillin (pUC19).<br> |
| - | <b>Follow-up:</b> Inoculated 5mL LB media (tetracycline positive) with cells from the transformation plates and incubated at 37<sup>o</sup>C overnight.<br> | + | <b>Follow-up:</b> Inoculated 5mL LB media (tetracycline positive) with cells from the transformation plates and incubated at 37<sup>o</sup>C overnight. (June 5/2010).<br><br> |
| - | <b>Objective:</ | + | <b>Objective:</b> Ligate pBad-TetR part with fluorescent protein part in pSB1C3 backbone.<br> |
| + | <b>Method:</b><br> | ||
| + | <u>Restriction</u><br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Name</b></td><td><b>Volume<br>pDNA (µL)</b></td><td><b>Volume<br>Water (µL)</b></td><td><b>Volume<br>Buffer (µL)</b></td><td><b>Enzymes</b></td><td><b>Total Volume</b></td></tr> | ||
| + | <tr><td>pSB-NEYFP (B4)</td><td>.8</td><td>43.7</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>pSB-CEYFP (B5)</td><td>.9</td><td>43.6</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>pBad-TetR)</td><td>.3</td><td>44.2</td><td>5</td><td>0.25µL EcoRI<br>0.25µL SpeI</td><td>50</td></tr> | ||
| + | <tr><td>NEYFP (E1)</td><td>4.3</td><td>40.7</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>NEYFP (E2)</td><td>0.3</td><td>42.2</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>Fusion CEYFP (E3)</td><td>3.9</td><td>40.6</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>Fusion CEYFP (E4)</td><td>2.0</td><td>42.5</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>Fusion CEYFP (E5)</td><td>3.0</td><td>41.5</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>CEYFP (E6)</td><td>0.6</td><td>43.9</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>CEYFP (E7)</td><td>0.5</td><td>44.0</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>pBad-TetR (F4)</td><td>2.5</td><td>42</td><td>5</td><td>0.25µL EcoRI<br>0.25µL SpeI</td><td>50</td></tr> | ||
| + | <tr><td>pBad-TetR (F5)</td><td>1.7</td><td>42.8</td><td>5</td><td>0.25µL EcoRI<br>0.25µL SpeI</td><td>50</td></tr> | ||
| + | <tr><td>pSB-CEYFP (G4)</td><td>2.9</td><td>41.6</td><td>5</td><td>0.25µL XbaI<br>0.25µL PstI</td><td>50</td></tr> | ||
| + | <tr><td>pSB1C3</td><td>15.5</td><td>46</td><td></td><td>0.25µL EcoRI<br>0.25µL PstI</td><td>62</td></tr></table> | ||
| + | Incubated at 37<sup>o</sup>C for 75 minutes.<br> | ||
| + | |||
| + | *Used Red buffer for the EcoRI/SpeI and EcoRI/PstI digests | ||
| + | *Used Tango buffer for the XbaI/PstI digests | ||
| + | *Did not heat kill upon removal from incubation, put directly into -20<sup>o</sup>C fridge. | ||
| + | |||
| + | <b>Continue Ligation on Saturday (See below).</b><br> | ||
| + | |||
| + | ==<font color="white">June 5/2010 == | ||
| + | (In the lab:AS)<br> | ||
| + | <b>Objective:</b> Ligate restriction products from June 3/2010.<br> | ||
| + | <b>Relevant information:</b> <br> | ||
| + | *Have 3 tubes of part 1 (pBad-TetR) | ||
| + | **In ampicillin backbone | ||
| + | *Have 10 tubes of part 2 (fluorescent protein - various) | ||
| + | **In ampicillin backbone | ||
| + | *Will have 30 combinations | ||
| + | *Will use pSB1C3 as plasmid backbone | ||
| + | **Used most of the pSB1T3 and want to save remainder for creating new backbone via PCR. | ||
| + | <b>Method:</b><br> | ||
| + | <b>*Restriction digests were not heat killed after reactions. Freezing probably killed the restriction enzymes, but I will hea kill them at 80<sup>o</sup>C for 20 minutes anyways prior to adding Ligase.</b> | ||
| + | **Cool on ice for 10 minutes before adding ligase. | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Master Mix</b></td><td><b>Volume/tube (µL)</b></td><td><b>Total Volume (µL)</b></td></tr> | ||
| + | <tr><td>DNA</td><td>6</td><td>---</td></tr> | ||
| + | <tr><td>10x Buffer</td><td>1</td><td>32</td></tr> | ||
| + | <tr><td>T4 DNA Ligase</td><td>.25</td><td>8</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>2.75</td><td>88</td></tr></table> | ||
| + | *Add 4µL master mix to each DNA tube.<br> | ||
| + | <b>Follow-up:</b> Ligation reactions will be transformed into DH5α cells<br> | ||
| + | |||
| + | |||
| + | ==<font color="white">June 6/2010 == | ||
| + | (In Lab: JV, HS)<br> | ||
| - | |||
<b>Objective:</b><br> Isolate the following plasmid DNA from DH5α:<br> | <b>Objective:</b><br> Isolate the following plasmid DNA from DH5α:<br> | ||
| Line 236: | Line 448: | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| - | Followed [[Team:Lethbridge/Notebook/Protocols|boiling lysis miniprep]] protocol. Eluted with 10 Milli-Q H<sub>2</sub>O and RNase A.<br> | + | Followed [[Team:Lethbridge/Notebook/Protocols|boiling lysis miniprep]] protocol. Eluted with 10µL Milli-Q H<sub>2</sub>O and RNase A.<br> |
| - | ===June 3/2010=== | + | <b>Notes:</b><br> |
| + | *Placed colony 2 in cell E10 of glycerol stocks and J6 of working plasmid box. | ||
| + | *Placed colony 2 in cell F1 of glycerol stocks and J5 of working plasmid box. | ||
| + | |||
| + | <b>Objective:</b><br> Transformed the following plasmid DNA into DH5α cells:<br> | ||
| + | |||
| + | *pBad-TetR-CEYFP: (F5+E6), (B10+E7), (B10+E6), (F4+E6), (F5+E4), (F4+E7)<br> | ||
| + | *pBad-TetR-Fusion CEYFP: (F5+E3), (B10+E4), (F4+E3), (B10+E3), (B10+E5), (F4+E4), (F5+E4), (F5+E5), (F4+E5)<br> | ||
| + | *pBad-TetR-pSB CEYFP: (F4+B5), (B10+G4), (F4+G4), (F4+B5), (F5+B5), (F5+G4)<br> | ||
| + | *pBad-TetR-NEYFP: (F5+E1), (B10+E1), (F4+E2), (F5+E2), (B10+E2), (F4+E1)<br> | ||
| + | *pBad-TetR-pSB NEYFP: (F5+B4), (F4+B4), (B10+B4)<br> | ||
| + | |||
| + | *Positive control -> DH5α + pSB1C3<br> | ||
| + | *Negative control -> DH%α + Milli-Q H<sub>2</sub>O<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | Followed [[Team:Lethbridge/Notebook/Protocols|Competent Cell Transformation]] protocol and used chloramphenicol as an antibiotic. We plated all 200µL of DNA onto the plates. The plates were incubated at 37<sup>o</sup>C from 4:30pm to 10:00am. | ||
| + | |||
| + | <b>Results:</b><br> | ||
| + | None of the plates showed any growth. | ||
| + | |||
| + | ==<font color="white">June 7/2010 == | ||
| + | (In Lab: JV, HB, TF, AV)<br> | ||
| + | |||
| + | <b>Objective:</b><br> Purification of DNA to increase ligation and transformation efficiency.<br> | ||
| + | |||
| + | <b>Method:</b><br> BioBasic Protocol for Purification of PCR products.<br> | ||
| + | |||
| + | DNA (from Working Plasmid Box) Purified: | ||
| + | *pBAD-TetR (B10) | ||
| + | *pBAD- TetR (F4) | ||
| + | *pBAD-TetR (F5) | ||
| + | *EYFP (B1) | ||
| + | *pSB-NEYFP (B4) | ||
| + | *pSB-CEYFP (B5) | ||
| + | *NEYFP (E1) | ||
| + | *NEYFP (E2) | ||
| + | *Fusion CEYFP (E3) | ||
| + | *Fusion CEYFP (E4) | ||
| + | *Fusion CEYFP (E5) | ||
| + | *CEYFP (E6) | ||
| + | *CEYFP (E7) | ||
| + | *EYFP (E8) | ||
| + | *EYFP (E9) | ||
| + | *EYFP (E10) | ||
| + | *ECFP (F1) | ||
| + | *ECFP (F2) | ||
| + | *ECFP (F3) | ||
| + | *EYFP (G1) | ||
| + | *pSB-CEYFP (G4) | ||
| + | |||
| + | |||
| + | <b>Objective:</b><br> Restrict and run 1% agarose gel of plasmids J5, J6, and pSB1T3 (June 2/10).<br> | ||
| + | |||
| + | |||
| + | <b>Method:</b> | ||
| + | |||
| + | <b>Restrictions</b> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Component</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>Restriction Enzyme (XbaI)</td><td>0.25</td></tr> | ||
| + | <tr><td>Buffer (Tango)</td><td>2</td></tr> | ||
| + | <tr><td>Plasmid DNA</td><td>2</td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>15.75</td></tr> | ||
| + | </table> | ||
| + | |||
| + | Incubated for 1 hour in 37<sup>o</sup>C incubator.<br> | ||
| + | Heat shocked for 10 min at 65<sup>o</sup>C on heating block. | ||
| + | |||
| + | |||
| + | <b>1% Agarose Gel</b> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>1kb ladder</td><td>2 ladder + 2 dye (6X) + 8 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>J5-pLacI-SRBS-Lumazine-dT(1)</td><td>10 DNA + 2 Dye (6X)</td></tr> | ||
| + | <tr><td>3</td><td>J5-pLacI-SRBS-Lumazine-dT(1)restricted</td><td>10 DNA + 2 Dye (6X)</td></tr> | ||
| + | <tr><td>4</td><td>J6-pLacI-SRBS-Lumazine-dT(2)</td><td>10 DNA + 2 Dye (6X)</td></tr> | ||
| + | <tr><td>5</td><td>J5-pLacI-SRBS-Lumazine-dT(2)restricted</td><td>10 DNA + 2 Dye (6X)</td></tr> | ||
| + | <tr><td>6</td><td>pSB1T3</td><td>10 DNA + 2 Dye (6X)</td></tr> | ||
| + | <tr><td>7</td><td>pSB1T3 restricted</td><td>10 DNA + 2 Dye (6X)</td></tr> | ||
| + | <tr><td>8</td><td>Empty</td><td></td></tr> | ||
| + | <tr><td>9</td><td>Empty</td><td></td></tr> | ||
| + | <tr><td>10</td><td>Empty</td><td></td></tr></table> | ||
| + | |||
| + | Ran at 100V for 72 min.<br> | ||
| + | |||
| + | <font color ="Red">IMAGE TO COME!!!!!!</font><br> | ||
| + | |||
| + | ==<font color="white">June 7/2010 - Evening == | ||
| + | (In Lab: AS, KG)<br> | ||
| + | |||
| + | <b>Objective:</b><br> Ligation of:<br> | ||
| + | *pBAD-TetR (F5) + CEYFP (E6) | ||
| + | *pBAD-TetR (F5) + NEYFP (E1) | ||
| + | *pBAD-TetR (F5) + pSB-NEYFP (B4) | ||
| + | *pBAD-TetR (F5) + Fusion-CEYFP (E7) | ||
| + | *pBAD-TetR (F5) + Fusion-CEYFP (E3) | ||
| + | *pBAD-TetR (F5) + CEYFP (E7) | ||
| + | *pBAD-TetR (F5) + NEYFP (E2) | ||
| + | *pBAD-TetR (F5) + pSB-CEYFP (B5) | ||
| + | *pBAD-TetR (F5) + Fusion-CEYFP (E5) | ||
| + | *pBAD-TetR (F5) + pSB-CEYFP (G4) | ||
| + | *pBAD-TetR (F4) + CEYFP (E7) | ||
| + | *pBAD-TetR (F4) + NEYFP (E1) | ||
| + | *pBAD-TetR (F4) + pSB-NEYFP (B4) | ||
| + | *pBAD-TetR (F4) + pSB-CEYFP (B5) | ||
| + | *pBAD-TetR (F4) + CEYFP (E6) | ||
| + | *pBAD-TetR (F4) + Fusion-CEYFP (E4) | ||
| + | *pBAD-TetR (F4) + Fusion-CEYFP (E3) | ||
| + | *pBAD-TetR (F4) + pSB-CEYFP (G4) | ||
| + | *pBAD-TetR (F4) + Fusion-CEYFP (E5) | ||
| + | *pBAD-TetR (F4) + NEYFP (E2) | ||
| + | *pBAD-TetR (B10) + pSB-NEYFP (B4) | ||
| + | *pBAD-TetR (B10) + pSB-CEYFP (G4) | ||
| + | *pBAD-TetR (B10) + NEYFP (E1) | ||
| + | *pBAD-TetR (B10) + CEYFP (E6) | ||
| + | *pBAD-TetR (B10) + CEYFP (E7) | ||
| + | *pBAD-TetR (B10) + Fusion-CEYFP (E4) | ||
| + | *pBAD-TetR (B10) + pSB-CEYFP (B5) | ||
| + | *pBAD-TetR (B10) + Fusion-CEYFP (E5) | ||
| + | *pBAD-TetR (B10) + Fusion-CEYFP (E3) | ||
| + | *pBAD-TetR (B10) + NEYFP (E2) | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | <font color ="Red">FILL ME IN!!!!!!</font><br> | ||
| + | |||
| + | ==<font color="white">June 8/2010== | ||
| + | (In the lab: JV, AV)<br> | ||
| + | <b>Objective:</b> Follow the overexpression of our pLacI-sRBS-Lum-dT construct.<br> | ||
| + | <b>Method:</b> <font color ="Red">FILL ME OUT!!!!!!</font><br> | ||
| + | <b>Results:</b> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Time</b></td><td><b>OD<sub>600</sub> F1</b></td><td><b>OD<sub>600</sub> E10</b></td></tr> | ||
| + | <tr><td>0</td><td>0.118</td><td>0.103</td></tr> | ||
| + | <tr><td>30</td><td>0.133</td><td>0.111</td></tr> | ||
| + | <tr><td>30</td><td>0.145</td><td>0.116</td></tr> | ||
| + | <tr><td>90</td><td></td><td></td></tr> | ||
| + | <tr><td>120</td><td>0.160</td><td>0.124</td></tr> | ||
| + | <tr><td>150</td><td>0.120</td><td>0.093</td></tr> | ||
| + | <tr><td>180</td><td>0.129</td><td>0.100</td></tr> | ||
| + | <tr><td>210</td><td>0.145</td><td>0.122</td></tr> | ||
| + | <tr><td>240</td><td>0.158</td><td>0.145</td></tr> | ||
| + | <tr><td>270</td><td>0.171</td><td>0.178</td></tr> | ||
| + | <tr><td>300</td><td>0.194</td><td>0.222</td></tr> | ||
| + | <tr><td>330</td><td>0.223</td><td>0.280</td></tr> | ||
| + | <tr><td>360</td><td>0.252</td><td>0.364</td></tr> | ||
| + | <tr><td>390</td><td>0.296</td><td>0.458</td></tr> | ||
| + | <tr><td>420</td><td>0.338</td><td>0.557<sup>†</sup></td></tr> | ||
| + | <tr><td>450</td><td>0.394</td><td>0.656</td></tr> | ||
| + | <tr><td>480</td><td>0.453</td><td>0.675</td></tr> | ||
| + | <tr><td>510</td><td>0.530</td><td>0.688</td></tr> | ||
| + | <tr><td>540</td><td>0.598<sup>†</sup></td><td>0.706</td></tr> | ||
| + | <tr><td>600</td><td>0.633</td><td>0.752</td></tr> | ||
| + | <tr><td>660</td><td>0.653</td><td></td></tr> | ||
| + | <tr><td>720</td><td>0.679</td><td></td></tr> | ||
| + | <tr><td>∞</td><td>1.278</td><td></td></tr></table> | ||
| + | † Overexpression induced by adding 1mM IPTG.<br> | ||
| + | Following overexpression, 1mL of cells was removed from the culture, spun down at ~13000xg for 20 seconds, excess media removed and rinsed with water.<br> | ||
| + | Suspended cells in 8M urea, mixed with 6x dye and ran on 18% SDS-PAGE gel for 90 minutes at 200V. Gel stained overnight.<br> | ||
| + | <b>Results:</b> <font color ="Red">IMAGE TO COME!!!!</font><br><br> | ||
| + | |||
| + | <b>Objective:</b> Calculate quantity of DNA in pBad-TetR and fluorescent protein mini-preps by staining an agarose gel. <br> | ||
| + | <b>Method:</b> Restrict plasmid DNA (done by AV,HB,TF on June 7/2010) and run on a 1% TAE agarose gel (JV).<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Gel 1<br>Sample</b></td><td><b>Gel 1 Load</b></td><td><b>Gel 2<br>Sample</b></td><td><b>Gel 2 Load</b></td></tr> | ||
| + | <tr><td>1</td><td>1kb Ladder</td><td>2µL dye, 2µL ladder<br>8µL MilliQ H<sub>2</sub>O</td><td>1kb Ladder</td><td>2µL dye, 2µL ladder<br>8µL MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>Restricted<br>EYFP (B1)</td><td>10µL DNA<br>2µL Dye</td><td>Restricted<br>Fusion CEYFP (E3)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>3</td><td>Unrestricted<br>EYFP (B1)</td><td>10µL DNA<br>2µL Dye</td><td>Unrestricted<br>Fusion CEYFP (E3)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>4</td><td>Restricted<br>pSB-CEYFP (B5)</td><td>10µL DNA<br>2µL Dye</td><td>Restricted<br>Fusion CEYFP (E4)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>5</td><td>Unrestricted<br>pSB-CEYFP (B5)</td><td>10µL DNA<br>2µL Dye</td><td>Unrestricted<br>Fusion CEYFP (E4)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>6</td><td>Restricted<br>ECFP (F2)</td><td>10µL DNA<br>2µL Dye</td><td>Restricted<br>EYFP (E10)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>7</td><td>Unrestricted<br>ECFP (F2)</td><td>10µL DNA<br>2µL Dye</td><td>Unrestricted<br>EYFP (E10)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>8</td><td>Restricted<br>pSB-CEYFP (G4)</td><td>10µL DNA<br>2µL Dye</td><td>Restricted<br>pSB NEYFP (B4)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>9</td><td>Unrestricted<br>pSB-CEYFP (G4)</td><td>10µL DNA<br>2µL Dye</td><td>Unrestricted<br>pSB NEYFP (B4)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>10</td><td>Restricted<br>EYFP (G1)</td><td>10µL DNA<br>2µL Dye</td><td>Restricted<br>ECFP (F3)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>11</td><td>Unrestricted<br>EYFP (G1)</td><td>10µL DNA<br>2µL Dye</td><td>Unrestricted<br>ECFP (F3)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>12</td><td>Restricted<br>NEYFP (E2)</td><td>10µL DNA<br>2µL Dye</td><td>pBad-TetR (F5)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>13</td><td>Unrestricted<br>NEYFP (E2)</td><td>10µL DNA<br>2µL Dye</td><td>Restricted<br>Fusion CEYFP (E5)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>14</td><td>Restricted<br>pBad-TetR (B10)</td><td>10µL DNA<br>2µL Dye</td><td>Unrestricted<br>Fusion CEYFP (E5)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>15</td><td>Unrestricted<br>pBad-TetR (B10)</td><td>10µL DNA<br>2µL Dye</td><td>pBad-TetR (F4)</td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>16</td><td>Restricted<br>CEYFP (E6)</td><td>10µL DNA<br>2µL Dye</td><td>Restricted<br>pSB1T3 </td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>17</td><td>Unrestricted<br>CEYFP (E6)</td><td>10µL DNA<br>2µL Dye</td><td>Unrestricted pSB1T3 </td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>18</td><td>Restricted<br>NEYFP (E1)</td><td>10µL DNA<br>2µL Dye</td><td></td><td>10µL DNA<br>2µL Dye</td></tr> | ||
| + | <tr><td>19</td><td>Unrestricted<br>NEYFP (E1)</td><td>10µL DNA<br>2µL Dye</td><td></td><td></td></tr> | ||
| + | <tr><td>20</td><td>Restricted<br>ECFP (F1)</td><td>10µL DNA<br>2µL Dye</td><td></td><td></td></tr> | ||
| + | <tr><td>21</td><td>Unrestricted<br>ECFP (F1)</td><td>10µL DNA<br>2µL Dye</td><td></td><td></td></tr> | ||
| + | <tr><td>22</td><td>Restricted<br>EYFP (E9)</td><td>10µL DNA<br>2µL Dye</td><td></td><td></td></tr> | ||
| + | <tr><td>23</td><td>Unrestricted<br>EYFP (E9)</td><td>10µL DNA<br>2µL Dye</td><td></td><td></td></tr> | ||
| + | <tr><td>24</td><td>Restricted<br>CEYFP (E7)</td><td>10µL DNA<br>2µL Dye</td><td></td><td></td></tr> | ||
| + | <tr><td>25</td><td>Unrestricted<br>CEYFP (E7)</td><td>10µL DNA<br>2µL Dye</td><td></td><td></td></tr> | ||
| + | <tr><td>26</td><td>Restricted<br>EYFP (E8)</td><td>10µL DNA<br>2µL Dye</td><td></td><td></td></tr> | ||
| + | <tr><td>27</td><td>Unrestricted<br>EYFP (E8)</td><td>10µL DNA<br>2µL Dye</td><td></td><td></td></tr></table> | ||
| + | Ran gel at 100V for 90 minutes.<br> | ||
| + | <b>Results:</b><br> | ||
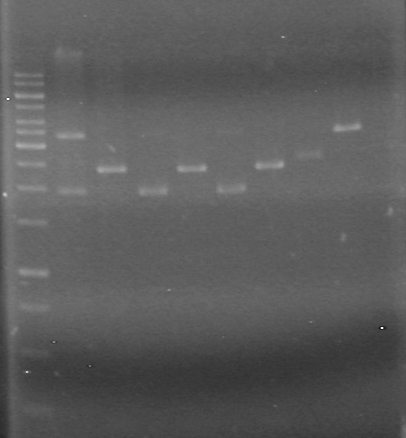
| + | [[image:100608JV.JPG|200px|none]] | ||
| + | No bands visible except for pSB1T3 lanes, therefore could not quantify anything that had not already been quantified.<br> | ||
| + | Also, purification of DNA done on June 7/2010 seemed to reduce amount of pDNA in sample.<br><br> | ||
| + | |||
| + | <b>Objective:</b> Restrict mms6 (D9,D10) and dT (C1) so we can ligate the dT onto the mms6 coding region.<br> | ||
| + | <b>Method:</b><br> | ||
| + | <u>mms6 Restriction</u><br> | ||
| + | *2µL mms6 pDNA | ||
| + | *2µL Red buffer | ||
| + | *0.25µL EcoRI | ||
| + | *0.25µL SpeI | ||
| + | *15.5µL MilliQ H<sub>2</sub>O | ||
| + | <u>dT Restriction</u><br> | ||
| + | *2µL dT pDNA | ||
| + | *2µL Orange buffer | ||
| + | *0.25µL EcoRI | ||
| + | *0.25µL XbaI | ||
| + | *15.5µL MilliQ H<sub>2</sub>O | ||
| + | |||
| + | Incubated for 1 hour at 37<sup>o</sup>C<br> | ||
| + | Heat shock on heat block (80<sup>o</sup>C) for 20 minutes<br> | ||
| + | <u>Ligation</u> | ||
| + | *2µL dT pDNA | ||
| + | *2µL mms6 pDNA | ||
| + | *0.25µL T4 DNA Ligase | ||
| + | *1µL 10x buffer | ||
| + | *4.75µL MilliQ H<sub>2</sub>O | ||
| + | |||
| + | Analyze results on 1% TAE agarose gel<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>1kb ladder</td><td>0.5 ladder; 2 dye; 9.5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>Unrestricted mms6 (D9)</td><td>10 DNA; 2 Dye</td></tr> | ||
| + | <tr><td>3</td><td>Restricted mms6 (D9)</td><td>10 DNA; 2 Dye</td></tr> | ||
| + | <tr><td>4</td><td>Unrestricted mms6 (D10)</td><td>10 DNA; 2 Dye</td></tr> | ||
| + | <tr><td>5</td><td>Restricted mms6 (D10)</td><td>10 DNA; 2 Dye</td></tr> | ||
| + | <tr><td>6</td><td>Unrestricted dT (C1)</td><td>10 DNA; 2 Dye</td></tr> | ||
| + | <tr><td>7</td><td>Restricted dT (C1)</td><td>10 DNA; 2 Dye</td></tr> | ||
| + | <tr><td>8</td><td>Empty</td><td></td></tr> | ||
| + | <tr><td>9</td><td>Empty</td><td></td></tr> | ||
| + | <tr><td>10</td><td>Empty</td><td></td></tr></table> | ||
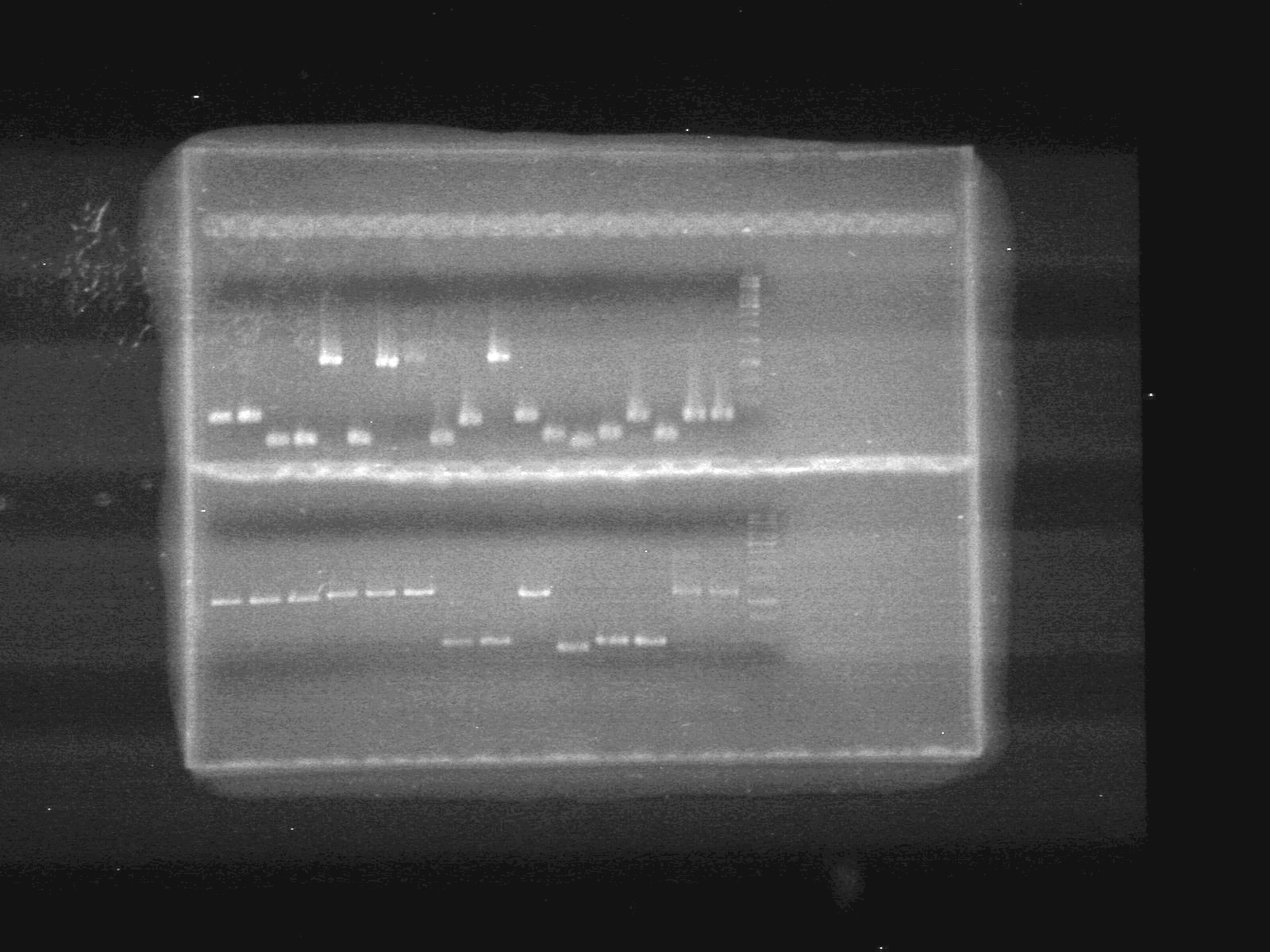
| + | <b>Results:</b><br> | ||
| + | [[image:100608-AV.AS.HS(3).jpg|200px|none]]<br> | ||
| + | Looks like the mms6 DNA was not cut at all. Therefore is doubtful that the ligations will work.<br><br> | ||
| + | |||
| + | ==<font color="white">June 8/2010 - Evening== | ||
| + | <b>Objective:</b> Transform pLacI-sRBS-Lum-dT constructs assembled via three antibiotic assembly on June 3/2010. Also transform mms6-dT constructs assembled today using old insertion method.<br> | ||
| + | <b>Method:</b> Follow [[Team:Lethbridge/Notebook/Protocols|transformation protocol]]. | ||
| + | <b>Results:</b> No colonies anywhere<br> | ||
| + | |||
| + | ==<font color="white">June 9/2010== | ||
| + | (in the lab: TF, JV)<br> | ||
| + | <b>Objective:</b> Transform mms6-dT ligation reactions from June 8/2010.<br> | ||
| + | <b>Method:</b> Followed [[Team:Lethbridge/Notebook/Protocols|transformation protocol]] and transformed the following: | ||
| + | *mms6 (D9) + dT (C1) | ||
| + | *mms6 (D10) + dT (C1) | ||
| + | *pUC19 (positive control) | ||
| + | *Water (negative control) | ||
| + | <b>Results:</b> No transformants on plates.<br><br> | ||
| + | |||
| + | ==<font color="white">June 10/2010== | ||
| + | (In the lab: AV, HB, JV)<br> | ||
| + | <b>Objective:</b> Repeat ligation of mms6 (B9,D9,D10) and dT (C1).<br> | ||
| + | <b>Method:</b><br> | ||
| + | <u>mms6 Restriction</u><br> | ||
| + | *2µL mms6 pDNA | ||
| + | *2µL Red buffer | ||
| + | *0.25µL EcoRI | ||
| + | *0.25µL SpeI | ||
| + | *15.5µL MilliQ H<sub>2</sub>O | ||
| + | <u>dT Restriction</u><br> | ||
| + | *2µL dT pDNA | ||
| + | *2µL Orange buffer | ||
| + | *0.25µL EcoRI | ||
| + | *0.25µL XbaI | ||
| + | *15.5µL MilliQ H<sub>2</sub>O | ||
| + | Incubated for 1 hour at 37<sup>o</sup>C.<br> | ||
| + | Killed enzymes by heating to 80<sup>o</sup>C for 20 minutes<br> | ||
| + | <u>Ligation</u> | ||
| + | *5µL dT pDNA | ||
| + | *5µL mms6 pDNA | ||
| + | *0.5µL T4 DNA Ligase | ||
| + | *2µL 10x buffer | ||
| + | *7.5µL MilliQ H<sub>2</sub>O | ||
| + | Incubated at room temperature overnight.<br> | ||
| + | <b>Results:</b><br> | ||
| + | Analyzed on 1% TAE agarose gel: | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>1kb ladder</td><td>0.5 ladder; 2 dye; 9.5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>Unrestricted mms6 (B9)</td><td>5 DNA; 2 dye; 5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>Restricted mms6 (B9)</td><td>5 DNA; 2 dye; 5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>4</td><td>Unrestricted mms6 (D9)</td><td>5 DNA; 2 dye; 5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>5</td><td>Restricted mms6 (D9)</td><td>5 DNA; 2 dye; 5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>6</td><td>Unrestricted mms6 (D10)</td><td>5 DNA; 2 dye; 5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>7</td><td>Restricted mms6 (D10)</td><td>5 DNA; 2 dye; 5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td>Unrestricted dT (C1)</td><td>5 DNA; 2 dye; 5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>9</td><td>Restricted dT (C1)</td><td>5 DNA; 2 dye; 5 MilliQ H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>10</td><td>D9 + C1 Ligation (June 8/2010)</td><td>5 DNA; 2 dye; 5 MilliQ H<sub>2</sub>O</td></tr></table> | ||
| + | Gel run for 80 minutes at 100V<br> | ||
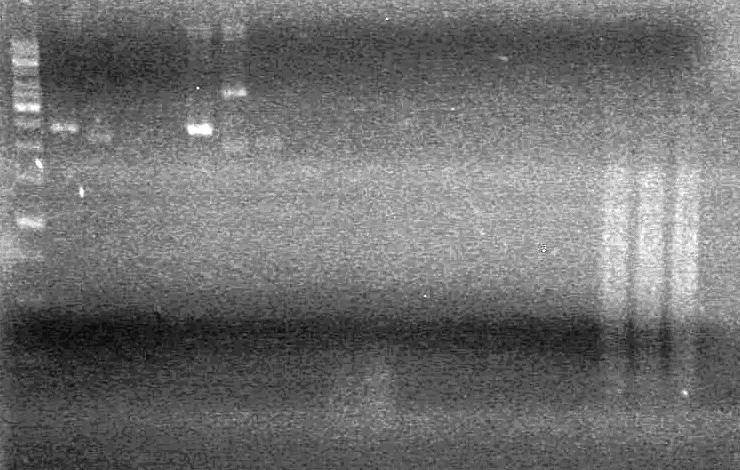
| + | [[image:100610-AV.HB.DM.JV.mms6 & DT.jpg|200px|none]]<br> | ||
| + | Looks like everything was restricted. Ligations may work this time. <br> | ||
| + | <b>Follow-up:</b> Ligation mixtures can be restriction tested and (if cutting works) transformed into DH5α cells.<br><br> | ||
| + | |||
| + | <b>Objective:</b> Transform pDNA from distribution plates into DH5α cells to generate glycerol stocks for lab.<br> | ||
| + | <b>Method:</b><br> | ||
| + | Remove DNA from the following locations on the 2010 distributions kits: | ||
| + | *double terminator (B0010-B0010; AKA B0017) from kit plate 2 well 6K (pSB1A2) | ||
| + | *double terminator (B0010-B0012; AKA B0015) from kit plate 1 well 23L (pSB1AK3) | ||
| + | Transform into DH5α cells using [[Team:Lethbridge/Notebook/Protocols|transformation protocol]], with pUC19 as positive control (1ng/µL), and water as negative control.<br> | ||
| + | <b>Results:</b><br> | ||
| + | *Positive control: TMTC (too many to count) colonies | ||
| + | *B0010-B0010: 41 colonies | ||
| + | *B0010-B0012: 120 colonies | ||
| + | |||
| + | ==<font color="white">June 14/2010 Evening== | ||
| + | (In the lab: TF, DM, HS)<br> | ||
| + | <b>Objective:</b> Restriction Digest of RBS-xylE with EcoRI and SpeI<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | Restriction Digestions<br> | ||
| + | *Pipetted 15.5µL of water, into 2 reaction tubes (0.6mL) each. | ||
| + | *Pipetted 2µL of red buffer into each of them. | ||
| + | *Pipetted 2µL of rbs-xylE plasmid DNA into both tubes, but taking only the thawed liquid on the side. | ||
| + | *Because this was not proper protocol for aliquoting DNA, we thawed the DNA out completely and then put 2µL more of the rbs-xylE plasmid DNA in. | ||
| + | *Pipetted 0.25µL of EcoRI enzyme and 0.25µL of SpeI enzyme into the restriction reaction tube. | ||
| + | *Placed in incubator (37<sup>o</sup>C) for 1 hour. | ||
| + | *Placed in ice for 10 minutes. | ||
| + | |||
| + | <b>Objective:</b> Test Ligations of rbs-xylE with T4-Ligase from iGEM -20<sup>o</sup>C and from HJ's lab's -20<sup>o</sup>C.<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | *Pipette 12.75µL of Milli-Q H<sub>2</sub>O into 4 mincrocentrifuge tubes. | ||
| + | *Pipette 2µL of 10X T4-Ligase buffer into all 4 tubes. | ||
| + | *Pipette 5µL of the RD product into both iGEM ligase tubes and HJ-Lab Ligase tubes. | ||
| + | *Pipetted 5µL of the RD control into both iGEM ligase tubes and HJ-Lab ligase tubes. | ||
| + | *Incubate overnight | ||
| + | |||
| + | ==<font color="white">June 15/2010== | ||
| + | (In the lab: JV)<br> | ||
| + | |||
| + | <b>Objective:</b>Continue T4-Ligase check and confirm that it is functional.<br> | ||
| + | |||
| + | <b>Method:</b>Run plasmid DNA;uncut, cut, and ligated on a 1% agarose gel (TAE)<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>rbs-xylE</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>2</td><td>rbs-xylE R.D.</td><td>10 DNA solution + 2 Dye<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>rbs-xylE [iGEM ligation]</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>4</td><td>rbs-xylE [HJ ligation]</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>5</td><td>rbs-xylE [iGEM ligation control]</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>6</td><td>rbs-xylE [HJ ligation control]</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>7</td><td>1kb ladder</td><td>2 ladder + 2 dye + 8 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td>mms6 (B9) R.D.</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>9</td><td>mms6 (B9)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>10</td><td>mms6 (D10) R.D.</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>11</td><td>mms6 (D10)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>12</td><td>mms6 (D9) R.D.</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>13</td><td>mms6 (D19)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>14</td><td>dt (C1) R.D.</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>15</td><td>dt (C1)</td><td>10 DNA solution + 2 Dye</td></tr></table> | ||
| + | |||
| + | |||
| + | <b>Result:</b>Ran gel for 89 minutes at 100V. Gel was unreadable. I will redo the gel using and 8 well comb. <font color ="Red">IMAGE TO COME!!!!</font><br> | ||
| + | |||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>rbs-xylE R.D.</td><td>5 DNA solution + 1 Dye</td></tr> | ||
| + | <tr><td>2</td><td>rbs-xylE</td><td>5 DNA solution + 1 Dye</td></tr> | ||
| + | <tr><td>3</td><td>iGEM ligation</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>4</td><td>iGEM ligation control</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>5</td><td>HJ ligation</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>6</td><td>HJ ligation control</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>7</td><td>1kb ladder</td><td>2 ladder + 2 dye + 8 Milli-Q H<sub>2</sub>O</td></tr></table> | ||
| + | |||
| + | Ran gel at 100V for 82 minutes.<br> | ||
| + | |||
| + | <b>Result:</b>The gel "broke" while running. Jeff hypothesized the cause to be excessive heat. However, the DNA was still visible.<font color ="Red">IMAGE TO COME!!!!</font><br> | ||
| + | |||
| + | <b>Objective:</b>Isolate plasmid DNA from DH5α cells.<br> | ||
| + | |||
| + | <b>Method:</b>The plasmid DNA is:<br> | ||
| + | * 1)double terminator B0017 (B0010-B0010) from 2010 kit plate 2:6K pSB1A2 | ||
| + | * 2)double terminator B0015 (B0010-B0012) from 2010 kit plate 1:23L pSB1AK2 | ||
| + | |||
| + | This will give us 2 additional dt's (3 total) into our working plasmid box to ligate onto the end of our constructs.<br> | ||
| + | |||
| + | Used the [[Team:Lethbridge/Notebook/Protocols|boiling lysis miniprep]] protocol and elute with 50µL of Milli-Q H<sub>2</sub>O with RNAse A. <br/> | ||
| + | |||
| + | These plasmids (labeled 1 & 2 above) are in:<br> | ||
| + | *working plasmid box: 1). J9 2). J10<br> | ||
| + | *glycerol sotck 2010 box: 1). F2 2). F3<br> | ||
| + | |||
| + | |||
| + | DNA was then purified using the protocol for purification of PCR products (BioBasic). DNA was eluted with 35µL of elution buffer.<br> | ||
| + | |||
| + | ==<font color="white">June 15/2010 Evening== | ||
| + | (In the lab: AS)<br> | ||
| + | |||
| + | Adam: not convinced that the rbs-xylE was cut properly last night. There doesn't appear to be and band where the insert should be.<br> | ||
| + | |||
| + | <b>Objective: </b>To confirm that EcoRI & SpeI are cutting (also PstI). Use rbs-xylE as test plasmid DNA.<br> | ||
| + | |||
| + | <b>Method: </b>Digest rbs-xylE with the following enzyme combinations:<br> | ||
| + | *EcoRI + SpeI (old [i.e. John's]) + Red Buffer | ||
| + | *EcoRI + SpeI (new) + Red Buffer | ||
| + | *EcoRI + PstI + Red Buffer | ||
| + | *EcoRi + Red Buffer | ||
| + | *SpeI (old) + Tango Buffer | ||
| + | *SpeI (new) + Tango Buffer | ||
| + | *PstI + Red Buffer | ||
| + | *No enzyme controls (Red & Tango Buffer) | ||
| + | |||
| + | Reaction mix as follows: <br> | ||
| + | *Milli-Q H<sub>2</sub>O 15.5µL | ||
| + | *10X Buffer 2µL | ||
| + | *pDNA 2µL | ||
| + | *Enzymes 0.25µL + 0.25µL<br> | ||
| + | |||
| + | Incubated at 37<sup>o</sup>C for 1 hour . Analyze on 1% TAE Agarose gel as follows:<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>No Enzyme Control (Tango)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>2</td><td>No Enzyme Control (Red)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>3</td><td>PstI</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>4</td><td>SpeI (old)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>5</td><td>SpeI (new)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>6</td><td>EcoRI</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>7</td><td>EcoRI + SpeI (old)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>8</td><td>EcoRI + SpeI (new)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>9</td><td>EcoRI + PstI</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>10</td><td>1kb ladder</td><td>0.5 ladder + 2 dye + 9.5 Milli-Q H<sub>2</sub>Oe</td></tr></table> | ||
| + | |||
| + | |||
| + | <b>Results: </b>Everything cuts exactly as it should. Continue with ligation and analysis. <font color ="Red">IMAGE TO COME!!!!</font><br> | ||
| + | |||
| + | ==<font color="white">June 16/2010== | ||
| + | (in lab: JV) | ||
| + | <b>Objective: </b>To miniprep Adam's overnight cultures of xylE and rbs. Completing this will give us a working stock of this plasmid.<br> | ||
| + | |||
| + | <b>Method: </b>Use the [[Team:Lethbridge/Notebook/Protocols|boiling lysis miniprep]] protocol and pufrify using BioBasic protocol for purification of PCR products.<br> | ||
| + | |||
| + | <b>Objective: </b>Transform pLacI-sRBS-lumazine synthase-dt into BL21(DE3).<br> | ||
| + | |||
| + | <b>Method:</b> | ||
| + | *Followed [[Team:Lethbridge/Notebook/Protocols|transformation protocol]] <br> | ||
| + | Changes to the protocol include:<br> | ||
| + | *Added 5µL DNA to 50µL BL21(DE3). | ||
| + | *Incubated in 500µL SOC instead of 250µL LB | ||
| + | *incubated for 90 minutes at 37<sup>o</sup>C at 4:30pm. | ||
| + | |||
| + | <b>Results:</b> There wasn't any growth after overnight incubation. Possibility of an antibiotic mix-up. | ||
| + | |||
| + | ==<font color="white">June 16/2010 Evening== | ||
| + | (in lab: ADS) | ||
| + | |||
| + | <b>Objective:</b> Continue with ligation test. Use restriction products to test T4 DNA ligase.<br> | ||
| + | |||
| + | <b>Method:</b> Have 10µL of DNA remaining for each restriction. Ligate together one of the single cut reactions, and ligate one of the double cut reactions. 4µL of each sample will be tested against each ligase.<br> | ||
| + | i.e.<br> | ||
| + | *EcoRI +SpeI (new) cut vs HJ's ligase | ||
| + | *EcoRI + SpeI (new) cut vs iGEM ligase | ||
| + | *SpeI (new) cut vs HJ's ligase | ||
| + | *SpeI (new) cut vs iGEM ligase<br> | ||
| + | |||
| + | Reaction Mixture:<br> | ||
| + | *DNA -> 4µL | ||
| + | *10X Buffer -> 2µL | ||
| + | *Milli-Q -> 13.5µL | ||
| + | *Ligase -> 0.5µL<br> | ||
| + | |||
| + | Prior to setting up reaction, heat kill restriction enzymes by heating to 80<sup>o</sup>C for 20 minutes and subsequently cooling on ice for 10 minutes.<br> | ||
| + | |||
| + | Ligations begun at 6:50pm<br> | ||
| + | |||
| + | Will run samples at 30 minutes reaction time and overnight. <br> | ||
| + | |||
| + | Analyze ligations in a 1% TAE Agarose gel<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>No Enzyme Control (from last night)</td><td>1 DNA solution + 1 Dye + 4 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>EcoRI + SpeI (old)(from last night)</td><td>1 DNA solution + 1 Dye + 4 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>EcoRI + SpeI (new) vs HJ's Ligase</td><td>5 DNA solution + 1 Dye</td></tr> | ||
| + | <tr><td>4</td><td>EcoRI + SpeI (new) vs iGEM ligase</td><td>5 DNA solution + 1 Dye</td></tr> | ||
| + | <tr><td>5</td><td>SpeI (new) vs HJ's Ligase</td><td>5 DNA solution + 1 Dye</td></tr> | ||
| + | <tr><td>6</td><td>SpeI (new) vs iGEM ligase</td><td>5 DNA solution + 1 Dye</td></tr> | ||
| + | <tr><td>7</td><td>SpeI (old)(from last night)</td><td>1 DNA solution + 1 Dye + 4 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td>1kb Ladder</td><td>0.25 Ladder + 4.75 H<sub>2</sub>O + 1 Dye</td></tr></table> | ||
| + | |||
| + | |||
| + | No observable ligation occurring 30 minutes with both ligases.<br> | ||
| + | |||
| + | <b>Objective:</b> Prepare DNA for sequencing<br> | ||
| + | |||
| + | <b>Method:</b> Need 20µL of DNA at 100ng/µL for each reaction. Need 20µL of 10µM primer for every 5 reactions.<br> | ||
| + | |||
| + | Plasmids that need to be sequenced:<br> | ||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Name</b></td><td><b>Concentration (ng/µL)</b></td><td><b>Dilution Factor</b></td><td><b>Final Concentration (ng/µL)</b></td></tr> | ||
| + | <tr><td>A8-CFP complete</td><td>1395</td><td>1/5</td><td>280</td></tr> | ||
| + | <tr><td>B4-pSB NEYFP</td><td>1105</td><td>1/5</td><td>240</td></tr> | ||
| + | <tr><td>B5-pSB CEYFP</td><td>1055</td><td>1/5</td><td>210</td></tr> | ||
| + | <tr><td>B6-CFP complete</td><td>1285</td><td>1/5</td><td>260</td></tr> | ||
| + | <tr><td>D7-xylE</td><td>1820</td><td>1/10</td><td>182</td></tr> | ||
| + | <tr><td>D8-xylE</td><td>420</td><td>1/2</td><td>210</td></tr> | ||
| + | <tr><td>E1-NEYFP</td><td>235</td><td>1/2</td><td>117.5</td></tr> | ||
| + | <tr><td>E2-NEYFP</td><td>2980</td><td>1/10</td><td>300</td></tr> | ||
| + | <tr><td>E3-Fusion CEYFP</td><td>255</td><td>1/2</td><td>122.5</td></tr> | ||
| + | <tr><td>E4-Fusion CEYFP</td><td>490</td><td>1/2</td><td>245</td></tr> | ||
| + | <tr><td>E5-Fusion CEYFP</td><td>335</td><td>1/2</td><td>335</td></tr> | ||
| + | <tr><td>E6-CEYFP</td><td>1605</td><td>1/10</td><td>160</td></tr> | ||
| + | <tr><td>E7-CEYFP</td><td>1930</td><td>1/10</td><td>190</td></tr> | ||
| + | <tr><td>G4-pSB CEYFP</td><td>340</td><td>1/4</td><td>85</td></tr> | ||
| + | <tr><td>G5-CFP complete</td><td>355</td><td>1.4</td><td>89</td></tr> | ||
| + | </table> | ||
| + | |||
| + | Total of 15 primers -> 30 reactions<br> | ||
| + | 60µL of each VF2 & VR primers (10µL) -> send 65µL | ||
| + | |||
| + | ==<font color="white">June 17/2010== | ||
| + | (in lab: JV, HB) | ||
| + | <b>Objective:</b>Run Agarose gel of overnight samples from June 16/10 to test T4 DNA ligase<br> | ||
| + | |||
| + | <b>Method:</b>Analyze ligation on a 1% TAE agarose gel.<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>No Enzyme Control (from June 15)</td><td>2 DNA solution + 2 Dye + 8 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>EcoRI + SpeI (old)(from June 15)</td><td>2 DNA solution + 2 Dye + 8 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>EcoRI + SpeI (new) vs HJ's Ligase</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>4</td><td>EcoRI + SpeI (new) vs iGEM ligase</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>5</td><td>SpeI (new) vs HJ's Ligase</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>6</td><td>SpeI (new) vs iGEM ligase</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>7</td><td>SpeI (old)(from last night)</td><td>10 DNA solution + 2 Dye</td></tr> | ||
| + | <tr><td>8</td><td>1kb Ladder</td><td>0.5 Ladder + 9.5 H<sub>2</sub>O + 2 Dye</td></tr></table> | ||
| + | |||
| + | Ran for 80 minutes at 100V. | ||
| + | |||
| + | <b>Results:</b><br> | ||
| + | <font color ="Red">IMAGE TO COME!!!!</font><br> | ||
| + | |||
| + | ==<font color="white">June 17/2010 Evening== | ||
| + | (in lab: JS, AS, KG)<br> | ||
| + | |||
| + | <b>Objective:</b>Do PCR to observe ligations<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Master Mix</b></td><td><b>Volume/tube (µL)</b></td><td><b>Total Volume (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>10.8</td><td>59.4</td></tr> | ||
| + | <tr><td>DNA</td><td>2</td><td>-------</td></tr> | ||
| + | <tr><td>5X Phusion Buffer</td><td>4</td><td>22</td></tr> | ||
| + | <tr><td>10µM dNTP's</td><td>.25</td><td>8</td></tr> | ||
| + | <tr><td>Forward Primer</td><td>1</td><td>5.5</td></tr> | ||
| + | <tr><td>Reverse Primer</td><td>1</td><td>5.5</td></tr> | ||
| + | <tr><td>Polymerase</td><td>0.2</td><td>1.1</td></tr></table> | ||
| + | |||
| + | |||
| + | Reaction Mixtures:<br> | ||
| + | *HJ Double | ||
| + | *iGEM Double | ||
| + | *HJ Single | ||
| + | *iGEM Single | ||
| + | *controls (no forward or reverse primer and no DNA)<br> | ||
| + | |||
| + | ==<font color="white">June 18/2010== | ||
| + | (in lab: JV)<br> | ||
| + | |||
| + | <b>Objective:</b>Observe the ligations from June 17/10 that were amplified using PCR.<br> | ||
| + | |||
| + | <b>Method:</b>Observe using 1% TAE agarose gel electrophoresis. Ran the gel for 80 minutes at 100V.<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>8</td><td>1kb Ladder</td><td>2 Ladder + 8 H<sub>2</sub>O + 2 Dye</td></tr> | ||
| + | <tr><td>2</td><td>control no DNA</td><td>20 PCR reaction + 4 6X Dye and used 12 of this solution</td></tr> | ||
| + | <tr><td>3</td><td>control no reverse primer</td><td>20 PCR reaction + 4 6X Dye and used 12 of this solution</td></tr> | ||
| + | <tr><td>4</td><td>control no forward primer</td><td>20 PCR reaction + 4 6X Dye and used 12 of this solution</td></tr> | ||
| + | <tr><td>5</td><td>HJ single</td><td>20 PCR reaction + 4 6X Dye and used 12 of this solution</td></tr> | ||
| + | <tr><td>6</td><td>iGEM single</td><td>20 PCR reaction + 4 6X Dye and used 12 of this solution</td></tr> | ||
| + | <tr><td>7</td><td>HJ double</td><td>20 PCR reaction + 4 6X Dye and used 12 of this solution</td></tr> | ||
| + | <tr><td>8</td><td>HJ double</td><td>20 PCR reaction + 4 6X Dye and used 12 of this solution</td></tr></table> | ||
| + | |||
| + | <b>Results:</b><font color ="Red">IMAGE TO COME!!!!</font><br> | ||
| + | <br> | ||
| + | |||
| + | ==<font color="white">June 21/2010== | ||
| + | (in lab: JV)<br> | ||
| + | |||
| + | <b>Objective:</b>Create more pSB1A3 plasmid.<br> | ||
| + | |||
| + | <b>Method:</b>Run a PCR of the plasmid.<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Master Mix</b></td><td><b>Volume/tube (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>10.8</tr> | ||
| + | <tr><td>DNA</td><td>2</td> | ||
| + | <tr><td>5X Phusion Buffer</td><td>4</td> | ||
| + | <tr><td>10µM dNTP's</td><td>.25</td> | ||
| + | <tr><td>Forward Primer</td><td>1</td> DIlute 10X -> 0.1 + 0.9 MilliQ H<sub>2</sub>O | ||
| + | <tr><td>Reverse Primer</td><td>1</td> DIlute 10X -> 0.1 + 0.9 MilliQ H<sub>2</sub>O | ||
| + | <tr><td>Polymerase</td><td>0.2</td></table> | ||
| + | |||
| + | Use ligtest setting. WIll run 36 cycles. | ||
| + | |||
| + | Use 3 controls: | ||
| + | *no forward primer | ||
| + | *no reverse primer | ||
| + | *no DNA | ||
| + | |||
| + | <b>Results:</b>PCR samples were run on a gel (1% 1XTAE) with the PCR samples from June 22/10. This gel was run on June 22/10<br> | ||
| + | |||
| + | ==<font color="white">June 22/2010== | ||
| + | (in lab: JV, HS)<br> | ||
| + | |||
| + | <b>Objective:</b>To overexpress pLacI-sRBS-lumazine synthase-dt in DH5α cells.<br> | ||
| + | |||
| + | <b>Method:</b>Incubate cells in 500mL LB w/Tet. At OD 0.6 (600λ) induce overexpression of lumazine synthase with IPTG (1µM). Take T<sub>o</sub>, T<sub>1</sub>, T<sub>2</sub>, T<sub>3</sub> readings after induced with IPTG. | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Time</b></td><td><b>OD F1 (600λ)</b></td><td><b>OD E10 (600λ)</b></td></tr> | ||
| + | <tr><td>0</td><td>0.078</td><td>0.072</td></tr> | ||
| + | <tr><td>60</td><td>0.122</td><td>0.110</td></tr> | ||
| + | <tr><td>90</td><td>0.128</td><td>0.125</td></tr> | ||
| + | <tr><td>120</td><td>0.140</td><td>0.138</td></tr> | ||
| + | <tr><td>150</td><td>0.149</td><td>0.157</td></tr> | ||
| + | <tr><td>180</td><td>0.169</td><td>0.183</td></tr> | ||
| + | <tr><td>210</td><td>0.205</td><td>0.230</td></tr> | ||
| + | <tr><td>240</td><td>0.243</td><td>0.276</td></tr> | ||
| + | <tr><td>270</td><td>0.278</td><td>0.323</td></tr> | ||
| + | <tr><td>300</td><td>0.317</td><td>0.398</td></tr> | ||
| + | <tr><td>330</td><td>0.394</td><td>0.428</td></tr> | ||
| + | <tr><td>360</td><td>0.394</td><td>0.480</td></tr> | ||
| + | <tr><td>390</td><td>0.441</td><td>0.553</td></tr> | ||
| + | <tr><td>400</td><td>----</td><td>0.594 (induced)</td></tr> | ||
| + | <tr><td>410</td><td>0.502</td><td>-----</td></tr> | ||
| + | <tr><td>450</td><td>0.580 (induced)</td><td>0.736</td></tr> | ||
| + | <tr><td>510</td><td>0.804</td><td>1.005</td></tr> | ||
| + | <tr><td>570</td><td>1.065</td><td>1.066</td></tr> | ||
| + | <tr><td>630</td><td>1.053</td><td>1.043</td></tr> | ||
| + | <tr><td>∞</td><td>0.055 (1/10 dilution)</td><td>0.078 (1/10 dilution)</td></tr> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | <b>Objective:</b>PCR amplify the following biobricks:<br> | ||
| + | *dt (23L) | ||
| + | *dt (6K) | ||
| + | *dt (C1) | ||
| + | *mms6 | ||
| + | *control (no DNA) | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Master Mix</b></td><td><b>Volume/tube (µL)</b></td><td><b>Total Volume (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>----</td><td>89.6</td></tr> | ||
| + | <tr><td>DNA</td><td>2</td><td>-------</td></tr> | ||
| + | <tr><td>5X Phusion Buffer</td><td>4</td><td>24</td></tr> | ||
| + | <tr><td>10µM dNTP's</td><td>.4</td><td>2.4</td></tr> | ||
| + | <tr><td>Forward Primer</td><td>0.2</td><td>1.2</td></tr> | ||
| + | <tr><td>Reverse Primer</td><td>0.2</td><td>1.2</td></tr> | ||
| + | <tr><td>Polymerase</td><td>0.2</td><td>1.2</td></tr></table><br> | ||
| + | |||
| + | 1.2% Agarose gel electrophoresis (1X TAE), used to analyze the above PCR's. 25mL of agarose solution was used in the small gel rig.<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>pSB1A3</td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>no DNA</td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>no reverse primer</td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>4</td><td>no forward primer</td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>5</td><td>1kb ladder</td><td>0.5 ladder + 2 dye + 9.5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>6</td><td>mms6</td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>7</td><td>dt (23L)</td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td>dt (6K)</td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>9</td><td>dt (C1)</td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>10</td><td>no DNA</td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr></table><br> | ||
| + | |||
| + | Ran the gel for 40 minutes at 100V.<br> | ||
| + | |||
| + | ==<font color="white">June 22/2010 Evening== | ||
| + | (in lab: KG, AS)<br> | ||
| + | |||
| + | <b>Objective:</b>To run a PCR of:<br> | ||
| + | *dt (J9,J10,C1) | ||
| + | *mms6 (B9,F10) | ||
| + | *rbs-xylE (I1,I2) | ||
| + | *xylE (D7, D8) | ||
| + | *pLacI-sRBS-lumazine synthase-dt (I8,I9) | ||
| + | *sRBS-lumazine synthase-dt (A1,A2,B7,B8,G2,G3) | ||
| + | *pLacI (A9,D1) | ||
| + | *pSB1A3<br> | ||
| + | so that we have more DNA for restrictions and ligations. <br> | ||
| + | |||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Master Mix 1</b></td><td><b>Volume/tube (µL)</b></td><td><b>Total Volume (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>10.8</td><td>226.8</td></tr> | ||
| + | <tr><td>DNA</td><td>2</td><td>4</td></tr> | ||
| + | <tr><td>5X Phusion HF Buffer</td><td>4</td><td>84</td></tr> | ||
| + | <tr><td>10µM dNTP's</td><td>1</td><td>21</td></tr> | ||
| + | <tr><td>Forward Primer (VF2)</td><td>1</td><td>21</td></tr> | ||
| + | <tr><td>Reverse Primer (VR)</td><td>1</td><td>21</td></tr> | ||
| + | <tr><td>Fimnzzymes polymerase</td><td>0.2</td><td>4.2</td></tr></table><br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Master Mix 2</b></td><td><b>Volume/tube (µL)</b></td><td><b>Total Volume (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>10.8</td><td>226.8</td></tr> | ||
| + | <tr><td>DNA</td><td>2</td><td>4</td></tr> | ||
| + | <tr><td>5X Econo Taq Buffer</td><td>4</td><td>84</td></tr> | ||
| + | <tr><td>10µM dNTP's</td><td>1</td><td>21</td></tr> | ||
| + | <tr><td>Forward Primer (VF2)</td><td>1</td><td>21</td></tr> | ||
| + | <tr><td>Reverse Primer (VR)</td><td>1</td><td>21</td></tr> | ||
| + | <tr><td>Imitrages polymerase (Econo Taq)</td><td>0.2</td><td>4.2</td></tr></table><br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Master Mix for pSB1A3</b></td><td><b>Volume/tube (µL)</b></td><td><b>Total Volume (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>10.8</td><td>31</td></tr> | ||
| + | <tr><td>DNA</td><td>1</td><td>20</td></tr> | ||
| + | <tr><td>5X Phusion Buffer</td><td>4</td><td>10</td></tr> | ||
| + | <tr><td>10µM dNTP's</td><td>1</td><td>2.5</td></tr> | ||
| + | <tr><td>Primer (SB-prep-2Ea)</td><td>1</td><td>2.5</td></tr> | ||
| + | <tr><td>Primer (SB-prep-3P)</td><td>1</td><td>2.5</td></tr> | ||
| + | <tr><td>Fimnzzymes polymerase</td><td>0.2</td><td>0.5</td></tr></table><br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Master Mix for pSB1A3</b></td><td><b>Volume/tube (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>10.8</td></tr> | ||
| + | <tr><td>DNA</td><td>2</td></tr> | ||
| + | <tr><td>5X Econo Taq Buffer</td><td>4</td></tr> | ||
| + | <tr><td>10µM dNTP's</td><td>1</td></tr> | ||
| + | <tr><td>Primer (SB-prep-2Ea)</td><td>1</td></tr> | ||
| + | <tr><td>Primer (SB-prep-3P)</td><td>1</td></tr> | ||
| + | <tr><td>Imitrages polymerase</td><td>0.2</td></tr></table><br> | ||
| + | |||
| + | ==<font color="white">June 23/2010== | ||
| + | (in lab: JV, HS)<br> | ||
| + | |||
| + | <b>Objective:</b>: To run the pcr products on an agarose gel.<br> | ||
| + | |||
| + | <b>Method:</b> Run on 1% agarose gel using the large gel rig.<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>1kb DNA Ladder</td><td>0.5 Ladder + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td><font color ="Blue">Phu-dt-J9</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td><font color ="Blue">Phu-dt-J10</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>4</td><td><font color ="Blue">Phu-dt-C1</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>5</td><td><font color ="Blue">Fus-mms6-B9</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>6</td><td><font color ="Blue">Fus-mms6-F10</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>7</td><td><font color ="Blue">Fus-rbs-xylE-I1</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td><font color ="Blue">Fus-rbs-xylE-I2</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>9</td><td><font color ="Blue">Fus-xylE-D7</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>10</td><td><font color ="Blue">Fus-xylE-D8</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>11</td><td><font color ="Blue">Fus-pLacI-sRBS-lum-dt I8</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>12</td><td><font color ="Blue">Fus-pLacI-sRBS-lum-dt I9</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>13</td><td><font color ="Blue">Fus-sRBS-lum-dt A1</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>14</td><td><font color ="Blue">Phu-sRBS-lum-dt A2</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>15</td><td><font color ="Blue">Phu-sRBS-lum-dt B7</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>16</td><td><font color ="Blue">Phu-sRBS-lum-dt B8</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>17</td><td><font color ="Blue">Phu-sRBS-lum-dt G2</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>18</td><td><font color ="Blue">Phu-sRBS-lum-dt G3</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>19</td><td><font color ="Blue">Phu-pLacI A9</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>20</td><td><font color ="Blue">Phu-pLacI D1</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>21</td><td><font color ="Red">dt J9</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>22</td><td><font color ="Red">dt J10</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>23</td><td><font color ="Red">dt C1</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>24</td><td><font color ="Red">mms6 B9</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>25</td><td><font color ="Red">mms6 F10</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>26</td><td><font color ="Red">rbs-xylE I1</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>27</td><td><font color ="Red">rbs-xylE I2</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>28</td><td><font color ="Red">xylE D7</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>29</td><td><font color ="Red">xylE D8</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>30</td><td><font color ="Red">pLacI-sRBS-lum-dt I8</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>31</td><td><font color ="Red">pLacI-sRBS-lum-dt I9</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>32</td><td><font color ="Red">sRBS-lum-dt A1</font></td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>33</td><td><font color ="Red">sRBS-lum-dt A2</font></td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>34</td><td><font color ="Red">sRBS-lum-dt B7</font></td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>35</td><td><font color ="Red">sRBS-lum-dt B8</font></td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>36</td><td><font color ="Red">sRBS-lum-dt G2</font></td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>37</td><td><font color ="Red">sRBS-lum-dt G3</font></td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>38</td><td><font color ="Red">sRBS-lum-dt A9</font></td><td>1 PCR sample + 2 Dye + 9 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>39</td><td><font color ="Red">sRBS-lum-dt D1</font></td><td>0.5 ladder + 2 dye + 9.5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>40</td><td><font color ="Red">pSB1A3 control</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>41</td><td><font color ="Red">pSB1A3</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>42</td><td><font color ="Blue">pSB1A3 control</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>43</td><td><font color ="Blue">pSB1A3</font></td><td>5 PCR sample + 2 Dye + 3 H<sub>2</sub>O</td></tr></table><br> | ||
| + | |||
| + | Ran gel for 45 minutes at 100V and stained in EtBr for 20 minutes and de-stained for 10 minutes. | ||
| + | |||
| + | *all Econe tubes are <font color ="Red">red</font> | ||
| + | *all Phusion tubes are <font color ="Blue">blue</font><br> | ||
| + | |||
| + | <b>Results:</b> | ||
| + | <font color="red">IMAGE!!!</font> | ||
| + | |||
| + | |||
| + | <b>Objective:</b> Adam put in overnight cultures on June 22. I want to extract the plasmid DNA.<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | *Put in overnight cultures at 8:30pm and taken out at 9:30am next morning. | ||
| + | *All cultures were DH5α in LB w/ Amp. | ||
| + | *Cultures were incubated at 37<sub>o</sub>C. | ||
| + | *Next morning cultures were "minipreped" using [[Team:Lethbridge/Notebook/Protocols|boiling lysis miniprep]] protocol.<br> | ||
| + | |||
| + | |||
| + | <b>Results:</b> Will view plasmid DNA extracted, on a 1% agarose gel (1X TAE) run at 100V for an hour.<br> | ||
| + | |||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>1kb Ladder</td><td>0.5 Ladder + 2 Dye (6X) + 9.5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>CFP complete 2010 box C8</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>Fusion CEYFP 2007 box H5</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>4</td><td>NEYFP 2007 box J6</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>5</td><td>Fusion CEYFP 2007 box J5</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>6</td><td>CFP complete 2010 box A10</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>7</td><td>pSB-NEYFP 2010 box B6 (C1?)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td>Fusion CEYFP 2007 box I5</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>9</td><td>xylE 2007 box B9</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>10</td><td>CEYFP 2007 box G6</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>11</td><td>CFP complete 2010 box A6</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>12</td><td>pSB-CEYFP 2010 box C5</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>13</td><td>pSB-NEYFP 2010 box B6 (C1?)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>14</td><td>pSB-CEYFP 2010 BOX C3</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>15</td><td>xylE 2007 box C4</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>16</td><td>CEYFP 2007 box H6</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>17</td><td>NEYFP 2007 box J6</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr></table><br> | ||
| + | |||
| + | <font color ="Red">Picture to come!!!!!!!!!</font><br> | ||
| + | |||
| + | ==<font color="white">June 24/2010== | ||
| + | (in lab: JV, HS, AV)<br> | ||
| + | |||
| + | <b>Objective:</b>: To run a PCR of biobrick parts from out working plasmid box.<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | <table border ="3"> | ||
| + | <tr><td><b>Master Mix</b></td><td><b>Volume/tube (µL)</b></td><td><b>Total Volume (µL)</b></td></tr> | ||
| + | <tr><td>MilliQ H<sub>2</sub>O</td><td>12.8</td><td>448</td></tr> | ||
| + | <tr><td>DNA</td><td>1</td><td>-------</td></tr> | ||
| + | <tr><td>5X Phusion Buffer</td><td>4</td><td>140</td></tr> | ||
| + | <tr><td>10µM dNTP's</td><td>1</td><td>35</td></tr> | ||
| + | <tr><td>Forward Primer</td><td>0.5</td><td>17.5</td></tr> | ||
| + | <tr><td>Reverse Primer</td><td>0.5</td><td>17.5</td></tr> | ||
| + | <tr><td>Polymerase</td><td>0.2</td><td>7</td></tr></table><br> | ||
| + | |||
| + | Ran for 25 cycles on lig25 setting. | ||
| + | |||
| + | <b>Objective:</b>: Restriction Digest, Run on agarose gel, and ligate PCR products from June 23/10.<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | Six tubes for restriction digest:<br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Part 1</b></td><td><b>Part 2</b></td></tr> | ||
| + | <tr><td>Lane 8 (I2)</td><td>Lane 4 (C1)</td></tr> | ||
| + | <tr><td>Lane 19 (A9)</td><td>Lane 8 (I2)</td></tr> | ||
| + | <tr><td>Lane 19 (A9)</td><td>Lane 8 (I2)</td></tr></table><br> | ||
| + | |||
| + | Restriction Digest Reaction Mixture:<br> | ||
| + | <table border ="3"> | ||
| + | <tr><td><b>Ingredient</b></td><td><b>Volume (µL)</b></td></tr> | ||
| + | <tr><td>Plasmid DNA</td><td>2</td></tr> | ||
| + | <tr><td>Enzyme</td><td>0.5</td></tr> | ||
| + | <tr><td>Buffer</td><td>2</td></tr> | ||
| + | <tr><td>Milli-Q H<sub>2</sub>O</td><td>15.5</td></tr></table><br> | ||
| + | Incubate at 37<sup>o</sup>C for 1 hour. Then heat kill the enzymes at 80<sup>o</sup>C for 20 minutes.<br> | ||
| + | |||
| + | 2% Agarose gel electrophoresis:<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>50bp Ladder</td><td>1 Ladder + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>Fus-RBS-xylE (I2); Lane 8</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>Fus-RBS-xylE (I2) restricted; Lane 8</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>4</td><td>Phu-dT (C1); Lane 4</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>5</td><td>Phu-dT (C1) restricted; Lane 4</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>6</td><td>Phu-pLacI (A9); Lane 19(1)</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>7</td><td>Phu-pLacI (A9) restricted; Lane 19(1)</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td>Phu-pLacI (A9); Lane 19(2)</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>9</td><td>Phu-pLacI (A9) restricted; Lane 19(2)</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>10</td><td>Fus-RBS-xylE (I2); Lane 8(1)</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>11</td><td>Fus-RBS-xylE (I2) restricted; Lane 8(1)</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>12</td><td>Fus-RBS-xylE (I2); Lane 8(2)</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>13</td><td>Fus-RBS-xylE (I2) restricted; Lane 8(2)</td><td>5 PCR sample + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr></table><br> | ||
| + | |||
| + | Ran gel at 100V for 60 min and stained in EtBr for 15 min. | ||
| + | |||
| + | <font color ="Red">Picture to come!!!!!!!!!</font><br> | ||
| + | |||
| + | |||
| + | <b>Objective:</b>: Run 1% agarose gel of PCR samples from June 24/10.<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>A4-pBAD</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>A4-pBAD</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>A6-SRBS</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>4</td><td>A7-SRBS</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>5</td><td>A8-CFP Complete</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>6</td><td>A10-SRBS</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>7</td><td>B1-EYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td>B2-N-term tag</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>9</td><td>B4-pSB-NEYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>10</td><td>B5-pSB-NEYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>11</td><td>B6-CFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>12</td><td>B10-pBAD-TetR</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>13</td><td>D3</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>14</td><td>D4-C-term tag</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>15</td><td>D5-C-term tag</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>16</td><td>D6-PLacI</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>17</td><td>E2-NEYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>18</td><td>E6-CEYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>19</td><td>E7-CEYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>20</td><td>1kb ladder</td><td>0.5 ladder + 2 Dye (6X) + 9.5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>21</td><td>E8-EYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>22</td><td>E9-EYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>23</td><td>E10-ECFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>24</td><td>F1-ECFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>25</td><td>F2-ECFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>26</td><td>F3-ECFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>27</td><td>F4-pBAD-TetR</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>28</td><td>F5-pBAD-TetR</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>29</td><td>G1-EYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>30</td><td>G4-pSB-CEYFP</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>31</td><td>G6-pBAD(1)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>32</td><td>G7-pBAD(2)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>33</td><td>G8-N-term tag</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>33</td><td>G9-Lumazine</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>33</td><td>1kb ladder</td><td>0.5 ladder + 2 Dye (6X) + 9.5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | </table><br> | ||
| + | <b>Results:</b> [[image:Lethbridge_100624PCR.JPG|200px]] | ||
| + | |||
| + | ==<font color="white">June 28/2010== | ||
| + | (in lab: JV, HS, AV, HB)<br> | ||
| + | |||
| + | <b>Objective:</b>: To restrict, run 1% agarose gel, and ligate all mms6, lumazine, xylE into pET28(a) for overexpression tests.<br> | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | 1) Restrict all parts and pET28(a) with NotI<br> | ||
| + | |||
| + | 2) Run on agarose gel (1%)<br> | ||
| + | |||
| + | 3) Ligate parts into pET28(a)<br> | ||
| + | |||
| + | Parts: <br> | ||
| + | :from working plasmid box 2010:<br> | ||
| + | *A3-Lumazine | ||
| + | *B9-Mms6 | ||
| + | *D7-xylE | ||
| + | *D8-xylE | ||
| + | *I10-Mms6 | ||
| + | :from pink tray:<br> | ||
| + | *B9-xylE | ||
| + | *C4-xylE | ||
| + | *G9-Lumazine | ||
| + | :(2X) pET28(a)<br> | ||
| + | |||
| + | Restrictions:<br> | ||
| + | *2µL DNA | ||
| + | *2µL Orange Buffer | ||
| + | *0.25µL Enzyme (NotI) | ||
| + | *15.75µL Milli-Q H<sub>2</sub>O | ||
| + | |||
| + | Protocol:<br> | ||
| + | *1) Incubate for 1 hour at 37<sup>o</sup>C | ||
| + | *2) Heat kill for 20 minutes at 80<sup>o</sup>C (heat block)<br> | ||
| + | |||
| + | Agarose Gel:<br> | ||
| + | |||
| + | <table><table border ="3"> | ||
| + | <tr><td><b>Lane</b></td><td><b>Sample</b></td><td><b>Load (µL)</b></td></tr> | ||
| + | <tr><td>1</td><td>1kb ladder</td><td>0.5 ladder + 2 Dye (6X) + 9.5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>2</td><td>Lumazine A3 RD</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>3</td><td>Lumazine A3</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>4</td><td>Mms6 I10 RD</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>5</td><td>Mms6 I10</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>6</td><td>Mms6 B9 RD</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>7</td><td>Mms6 B9</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>8</td><td>xylE D8 RD</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>9</td><td>xylE D8</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>10</td><td>xylE D7 RD</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>11</td><td>xylE D7</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>12</td><td>Lumazine G9 RD (p)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>13</td><td>Lumazine G9 (p)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>14</td><td>xylE B9 RD (p)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>15</td><td>xylE B9 (p)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>16</td><td>xylE C4 RD (p)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>17</td><td>xylE C4 (p)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>18</td><td>pET28(a) RD</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>19</td><td>pET28(a) RD</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr> | ||
| + | <tr><td>20</td><td>pET28(a)</td><td>5 DNA + 2 Dye (6X) + 5 Milli-Q H<sub>2</sub>O</td></tr></table> | ||
| + | |||
| + | Ran gel at 100V for 90 minutes<br> | ||
| + | |||
| + | <b>Results:</b>pET28(a) bands are all smeared, and are not useful from inserting parts.<br> | ||
| + | [[image:Lethbridge_100628AV (2).JPG|200px]]<br> | ||
| + | |||
| + | ==<font color="white">June 30/2010== | ||
| + | (in lab: JV)<br> | ||
| + | |||
| + | <b>Objective:</b>: Isolate plasmid DNA from DH5α<br> | ||
| + | |||
| + | <b>Method:</b>Used Kothe Maxiprep protocol.<br> | ||
| + | Cell Pellet Weights:<br> | ||
| + | *xylE = 1.15g | ||
| + | *Mms6 = 1.13g | ||
| + | *pET28(a) = 0.67g | ||
| + | *lumazine synthase = 1.1g<br> | ||
| + | |||
| + | <b>Results:</b><br> | ||
 "
"