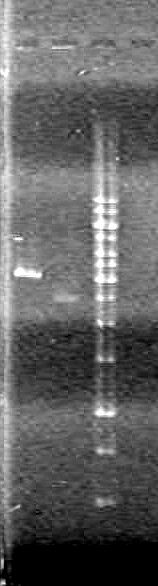
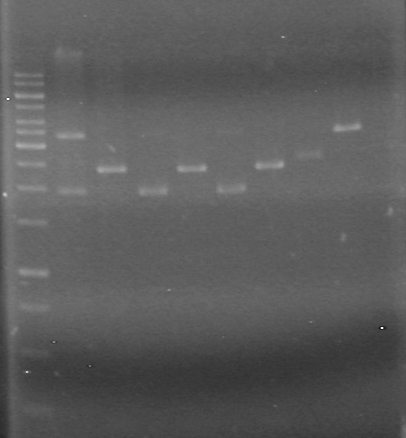
| Lane | Sample | Load (µL) |
| 1 | rbs-xylE R.D. | 5 DNA solution + 1 Dye |
| 2 | rbs-xylE | 5 DNA solution + 1 Dye |
| 3 | iGEM ligation | 10 DNA solution + 2 Dye |
| 4 | iGEM ligation control | 10 DNA solution + 2 Dye |
| 5 | HJ ligation | 10 DNA solution + 2 Dye |
| 6 | HJ ligation control | 10 DNA solution + 2 Dye |
| 7 | 1kb ladder | 2 ladder + 2 dye + 8 Milli-Q H2O |
Ran gel at 100V for 82 minutes.
Result:The gel "broke" while running. Jeff hypothesized the cause to be excessive heat. However, the DNA was still visible.IMAGE TO COME!!!!
Objective:Isolate plasmid DNA from DH5α cells.
Method:The plasmid DNA is:
- 1)double terminator B0017 (B0010-B0010) from 2010 kit plate 2:6K pSB1A2
- 2)double terminator B0015 (B0010-B0012) from 2010 kit plate 1:23L pSB1AK2
This will give us 2 additional dt's (3 total) into our working plasmid box to ligate onto the end of our constructs.
Used the boiling lysis miniprep protocol and elute with 50µL of Milli-Q H2O with RNAse A.
These plasmids (labeled 1 & 2 above) are in:
- working plasmid box: 1). J9 2). J10
- glycerol sotck 2010 box: 1). F2 2). F3
DNA was then purified using the protocol for purification of PCR products (BioBasic). DNA was eluted with 35µL of elution buffer.
June 15/2010 Evening
(In the lab: AS)
Adam: not convinced that the rbs-xylE was cut properly last night. There doesn't appear to be and band where the insert should be.
Objective: To confirm that EcoRI & SpeI are cutting (also PstI). Use rbs-xylE as test plasmid DNA.
Method: Digest rbs-xylE with the following enzyme combinations:
- EcoRI + SpeI (old [i.e. John's]) + Red Buffer
- EcoRI + SpeI (new) + Red Buffer
- EcoRI + PstI + Red Buffer
- EcoRi + Red Buffer
- SpeI (old) + Tango Buffer
- SpeI (new) + Tango Buffer
- PstI + Red Buffer
- No enzyme controls (Red & Tango Buffer)
Reaction mix as follows:
- Milli-Q H2O 15.5µL
- 10X Buffer 2µL
- pDNA 2µL
- Enzymes 0.25µL + 0.25µL
Incubated at 37oC for 1 hour . Analyze on 1% TAE Agarose gel as follows:
| Lane | Sample | Load (µL) |
| 1 | No Enzyme Control (Tango) | 10 DNA solution + 2 Dye |
| 2 | No Enzyme Control (Red) | 10 DNA solution + 2 Dye |
| 3 | PstI | 10 DNA solution + 2 Dye |
| 4 | SpeI (old) | 10 DNA solution + 2 Dye |
| 5 | SpeI (new) | 10 DNA solution + 2 Dye |
| 6 | EcoRI | 10 DNA solution + 2 Dye |
| 7 | EcoRI + SpeI (old) | 10 DNA solution + 2 Dye |
| 8 | EcoRI + SpeI (new) | 10 DNA solution + 2 Dye |
| 9 | EcoRI + PstI | 10 DNA solution + 2 Dye |
| 10 | 1kb ladder | 0.5 ladder + 2 dye + 9.5 Milli-Q H2Oe |
Results: Everything cuts exactly as it should. Continue with ligation and analysis. IMAGE TO COME!!!!
June 16/2010
(in lab: JV)
Objective: To miniprep Adam's overnight cultures of xylE and rbs. Completing this will give us a working stock of this plasmid.
Method: Use the boiling lysis miniprep protocol and pufrify using BioBasic protocol for purification of PCR products.
Objective: Transform pLacI-sRBS-lumazine synthase-dt into BL21(DE3).
Method:
Changes to the protocol include:
- Added 5µL DNA to 50µL BL21(DE3).
- Incubated in 500µL SOC instead of 250µL LB
- incubated for 90 minutes at 37oC at 4:30pm.
Results: There wasn't any growth after overnight incubation. Possibility of an antibiotic mix-up.
June 16/2010 Evening
(in lab: ADS)
Objective: Continue with ligation test. Use restriction products to test T4 DNA ligase.
Method: Have 10µL of DNA remaining for each restriction. Ligate together one of the single cut reactions, and ligate one of the double cut reactions. 4µL of each sample will be tested against each ligase.
i.e.
- EcoRI +SpeI (new) cut vs HJ's ligase
- EcoRI + SpeI (new) cut vs iGEM ligase
- SpeI (new) cut vs HJ's ligase
- SpeI (new) cut vs iGEM ligase
Reaction Mixture:
- DNA -> 4µL
- 10X Buffer -> 2µL
- Milli-Q -> 13.5µL
- Ligase -> 0.5µL
Prior to setting up reaction, heat kill restriction enzymes by heating to 80oC for 20 minutes and subsequently cooling on ice for 10 minutes.
Ligations begun at 6:50pm
Will run samples at 30 minutes reaction time and overnight.
Analyze ligations in a 1% TAE Agarose gel
| Lane | Sample | Load (µL) |
| 1 | No Enzyme Control (from last night) | 1 DNA solution + 1 Dye + 4 H2O |
| 2 | EcoRI + SpeI (old)(from last night) | 1 DNA solution + 1 Dye + 4 H2O |
| 3 | EcoRI + SpeI (new) vs HJ's Ligase | 5 DNA solution + 1 Dye |
| 4 | EcoRI + SpeI (new) vs iGEM ligase | 5 DNA solution + 1 Dye |
| 5 | SpeI (new) vs HJ's Ligase | 5 DNA solution + 1 Dye |
| 6 | SpeI (new) vs iGEM ligase | 5 DNA solution + 1 Dye |
| 7 | SpeI (old)(from last night) | 1 DNA solution + 1 Dye + 4 H2O |
| 8 | 1kb Ladder | 0.25 Ladder + 4.75 H2O + 1 Dye |
No observable ligation occurring 30 minutes with both ligases.
Objective: Prepare DNA for sequencing
Method: Need 20µL of DNA at 100ng/µL for each reaction. Need 20µL of 10µM primer for every 5 reactions.
Plasmids that need to be sequenced:
| Name | Concentration (ng/µL) | Dilution Factor | Final Concentration (ng/µL) |
| A8-CFP complete | 1395 | 1/5 | 280 |
| B4-pSB NEYFP | 1105 | 1/5 | 240 |
| B5-pSB CEYFP | 1055 | 1/5 | 210 |
| B6-CFP complete | 1285 | 1/5 | 260 |
| D7-xylE | 1820 | 1/10 | 182 |
| D8-xylE | 420 | 1/2 | 210 |
| E1-NEYFP | 235 | 1/2 | 117.5 |
| E2-NEYFP | 2980 | 1/10 | 300 |
| E3-Fusion CEYFP | 255 | 1/2 | 122.5 |
| E4-Fusion CEYFP | 490 | 1/2 | 245 |
| E5-Fusion CEYFP | 335 | 1/2 | 335 |
| E6-CEYFP | 1605 | 1/10 | 160 |
| E7-CEYFP | 1930 | 1/10 | 190 |
| G4-pSB CEYFP | 340 | 1/4 | 85 |
| G5-CFP complete | 355 | 1.4 | 89 |
Total of 15 primers -> 30 reactions
60µL of each VF2 & VR primers (10µL) -> send 65µL
June 17/2010
(in lab: JV, HB)
Objective:Run Agarose gel of overnight samples from June 16/10 to test T4 DNA ligase
Method:Analyze ligation on a 1% TAE agarose gel.
| Lane | Sample | Load (µL) |
| 1 | No Enzyme Control (from June 15) | 2 DNA solution + 2 Dye + 8 H2O |
| 2 | EcoRI + SpeI (old)(from June 15) | 2 DNA solution + 2 Dye + 8 H2O |
| 3 | EcoRI + SpeI (new) vs HJ's Ligase | 10 DNA solution + 2 Dye |
| 4 | EcoRI + SpeI (new) vs iGEM ligase | 10 DNA solution + 2 Dye |
| 5 | SpeI (new) vs HJ's Ligase | 10 DNA solution + 2 Dye |
| 6 | SpeI (new) vs iGEM ligase | 10 DNA solution + 2 Dye |
| 7 | SpeI (old)(from last night) | 10 DNA solution + 2 Dye |
| 8 | 1kb Ladder | 0.5 Ladder + 9.5 H2O + 2 Dye |
Ran for 80 minutes at 100V.
Results:
IMAGE TO COME!!!!
June 17/2010 Evening
(in lab: JS, AS, KG)
Objective:Do PCR to observe ligations
Method:
| Master Mix | Volume/tube (µL) | Total Volume (µL) |
| MilliQ H2O | 10.8 | 59.4 |
| DNA | 2 | ------- |
| 5X Phusion Buffer | 4 | 22 |
| 10µM dNTP's | .25 | 8 |
| Forward Primer | 1 | 5.5 |
| Reverse Primer | 1 | 5.5 |
| Polymerase | 0.2 | 1.1 |
Reaction Mixtures:
- HJ Double
- iGEM Double
- HJ Single
- iGEM Single
- controls (no forward or reverse primer and no DNA)
June 18/2010
(in lab: JV)
Objective:Observe the ligations from June 17/10 that were amplified using PCR.
Method:Observe using 1% TAE agarose gel electrophoresis. Ran the gel for 80 minutes at 100V.
| Lane | Sample | Volume (µL) |
| 8 | 1kb Ladder | 2 Ladder + 8 H2O + 2 Dye |
| 2 | control no DNA | 20 PCR reaction + 4 6X Dye and used 12 of this solution |
| 3 | control no reverse primer | 20 PCR reaction + 4 6X Dye and used 12 of this solution |
| 4 | control no forward primer | 20 PCR reaction + 4 6X Dye and used 12 of this solution |
| 5 | HJ single | 20 PCR reaction + 4 6X Dye and used 12 of this solution |
| 6 | iGEM single | 20 PCR reaction + 4 6X Dye and used 12 of this solution |
| 7 | HJ double | 20 PCR reaction + 4 6X Dye and used 12 of this solution |
| 8 | HJ double | 20 PCR reaction + 4 6X Dye and used 12 of this solution |
Results:IMAGE TO COME!!!!
June 21/2010
(in lab: JV)
Objective:Create more pSB1A3 plasmid.
Method:Run a PCR of the plasmid.
| Master Mix | Volume/tube (µL) |
| MilliQ H2O | 10.8 |
| DNA | 2 |
| 5X Phusion Buffer | 4 |
| 10µM dNTP's | .25 |
| Forward Primer | 1 | DIlute 10X -> 0.1 + 0.9 MilliQ H2O
| Reverse Primer | 1 | DIlute 10X -> 0.1 + 0.9 MilliQ H2O
| Polymerase | 0.2 |
Use ligtest setting. WIll run 36 cycles.
Use 3 controls:
- no forward primer
- no reverse primer
- no DNA
Results:PCR samples were run on a gel (1% 1XTAE) with the PCR samples from June 22/10. This gel was run on June 22/10
June 22/2010
(in lab: JV, HS)
Objective:To overexpress pLacI-sRBS-lumazine synthase-dt in DH5α cells.
Method:Incubate cells in 500mL LB w/Tet. At OD 0.6 (600λ) induce overexpression of lumazine synthase with IPTG (1µM). Take To, T1, T2, T3 readings after induced with IPTG.
| Time | OD F1 (600λ) | OD E10 (600λ) |
| 0 | 0.078 | 0.072 |
| 60 | 0.122 | 0.110 |
| 90 | 0.128 | 0.125 |
| 120 | 0.140 | 0.138 |
| 150 | 0.149 | 0.157 |
| 180 | 0.169 | 0.183 |
| 210 | 0.205 | 0.230 |
| 240 | 0.243 | 0.276 |
| 270 | 0.278 | 0.323 |
| 300 | 0.317 | 0.398 |
| 330 | 0.394 | 0.428 |
| 360 | 0.394 | 0.480 |
| 390 | 0.441 | 0.553 |
| 400 | ---- | 0.594 (induced) |
| 410 | 0.502 | ----- |
| 450 | 0.580 (induced) | 0.736 |
| 510 | 0.804 | 1.005 |
| 570 | 1.065 | 1.066 |
| 630 | 1.053 | 1.043 |
| ∞ | 0.055 (1/10 dilution) | 0.078 (1/10 dilution) |
Objective:PCR amplify the following biobricks:
- dt (23L)
- dt (6K)
- dt (C1)
- mms6
- control (no DNA)
Method:
| Master Mix | Volume/tube (µL) | Total Volume (µL) |
| MilliQ H2O | ---- | 89.6 |
| DNA | 2 | ------- |
| 5X Phusion Buffer | 4 | 24 |
| 10µM dNTP's | .4 | 2.4 |
| Forward Primer | 0.2 | 1.2 |
| Reverse Primer | 0.2 | 1.2 |
| Polymerase | 0.2 | 1.2 |
1.2% Agarose gel electrophoresis (1X TAE), used to analyze the above PCR's. 25mL of agarose solution was used in the small gel rig.
| Lane | Sample | Volume (µL) |
| 1 | pSB1A3 | 1 PCR sample + 2 Dye + 9 H2O |
| 2 | no DNA | 1 PCR sample + 2 Dye + 9 H2O |
| 3 | no reverse primer | 1 PCR sample + 2 Dye + 9 H2O |
| 4 | no forward primer | 1 PCR sample + 2 Dye + 9 H2O |
| 5 | 1kb ladder | 0.5 ladder + 2 dye + 9.5 Milli-Q H2O |
| 6 | mms6 | 1 PCR sample + 2 Dye + 9 H2O |
| 7 | dt (23L) | 1 PCR sample + 2 Dye + 9 H2O |
| 8 | dt (6K) | 1 PCR sample + 2 Dye + 9 H2O |
| 9 | dt (C1) | 1 PCR sample + 2 Dye + 9 H2O |
| 10 | no DNA | 1 PCR sample + 2 Dye + 9 H2O |
Ran the gel for 40 minutes at 100V.
June 22/2010 Evening
(in lab: KG, AS)
Objective:To run a PCR of:
- dt (J9,J10,C1)
- mms6 (B9,F10)
- rbs-xylE (I1,I2)
- xylE (D7, D8)
- pLacI-sRBS-lumazine synthase-dt (I8,I9)
- sRBS-lumazine synthase-dt (A1,A2,B7,B8,G2,G3)
- pLacI (A9,D1)
- pSB1A3
so that we have more DNA for restrictions and ligations.
Method:
| Master Mix 1 | Volume/tube (µL) | Total Volume (µL) |
| MilliQ H2O | 10.8 | 226.8 |
| DNA | 2 | 4 |
| 5X Phusion HF Buffer | 4 | 84 |
| 10µM dNTP's | 1 | 21 |
| Forward Primer (VF2) | 1 | 21 |
| Reverse Primer (VR) | 1 | 21 |
| Fimnzzymes polymerase | 0.2 | 4.2 |
| Master Mix 2 | Volume/tube (µL) | Total Volume (µL) |
| MilliQ H2O | 10.8 | 226.8 |
| DNA | 2 | 4 |
| 5X Econo Taq Buffer | 4 | 84 |
| 10µM dNTP's | 1 | 21 |
| Forward Primer (VF2) | 1 | 21 |
| Reverse Primer (VR) | 1 | 21 |
| Imitrages polymerase (Econo Taq) | 0.2 | 4.2 |
| Master Mix for pSB1A3 | Volume/tube (µL) | Total Volume (µL) |
| MilliQ H2O | 10.8 | 31 |
| DNA | 1 | 20 |
| 5X Phusion Buffer | 4 | 10 |
| 10µM dNTP's | 1 | 2.5 |
| Primer (SB-prep-2Ea) | 1 | 2.5 |
| Primer (SB-prep-3P) | 1 | 2.5 |
| Fimnzzymes polymerase | 0.2 | 0.5 |
| Master Mix for pSB1A3 | Volume/tube (µL) |
| MilliQ H2O | 10.8 |
| DNA | 2 |
| 5X Econo Taq Buffer | 4 |
| 10µM dNTP's | 1 |
| Primer (SB-prep-2Ea) | 1 |
| Primer (SB-prep-3P) | 1 |
| Imitrages polymerase | 0.2 |
June 23/2010
(in lab: JV, HS)
Objective:: To run the pcr products on an agarose gel.
Method: Run on 1% agarose gel using the large gel rig.
| Lane | Sample | Load (µL) |
| 1 | 1kb DNA Ladder | 0.5 Ladder + 2 Dye + 9 H2O |
| 2 | Phu-dt-J9 | 5 PCR sample + 2 Dye + 3 H2O |
| 3 | Phu-dt-J10 | 5 PCR sample + 2 Dye + 3 H2O |
| 4 | Phu-dt-C1 | 5 PCR sample + 2 Dye + 3 H2O |
| 5 | Fus-mms6-B9 | 5 PCR sample + 2 Dye + 3 H2O |
| 6 | Fus-mms6-F10 | 5 PCR sample + 2 Dye + 3 H2O |
| 7 | Fus-rbs-xylE-I1 | 5 PCR sample + 2 Dye + 3 H2O |
| 8 | Fus-rbs-xylE-I2 | 5 PCR sample + 2 Dye + 3 H2O |
| 9 | Fus-xylE-D7 | 5 PCR sample + 2 Dye + 3 H2O |
| 10 | Fus-xylE-D8 | 5 PCR sample + 2 Dye + 3 H2O |
| 11 | Fus-pLacI-sRBS-lum-dt I8 | 5 PCR sample + 2 Dye + 3 H2O |
| 12 | Fus-pLacI-sRBS-lum-dt I9 | 5 PCR sample + 2 Dye + 3 H2O |
| 13 | Fus-sRBS-lum-dt A1 | 5 PCR sample + 2 Dye + 3 H2O |
| 14 | Phu-sRBS-lum-dt A2 | 5 PCR sample + 2 Dye + 3 H2O |
| 15 | Phu-sRBS-lum-dt B7 | 5 PCR sample + 2 Dye + 3 H2O |
| 16 | Phu-sRBS-lum-dt B8 | 5 PCR sample + 2 Dye + 3 H2O |
| 17 | Phu-sRBS-lum-dt G2 | 5 PCR sample + 2 Dye + 3 H2O |
| 18 | Phu-sRBS-lum-dt G3 | 5 PCR sample + 2 Dye + 3 H2O |
| 19 | Phu-pLacI A9 | 5 PCR sample + 2 Dye + 3 H2O |
| 20 | Phu-pLacI D1 | 5 PCR sample + 2 Dye + 3 H2O |
| 21 | dt J9 | 5 PCR sample + 2 Dye + 3 H2O |
| 22 | dt J10 | 5 PCR sample + 2 Dye + 3 H2O |
| 23 | dt C1 | 5 PCR sample + 2 Dye + 3 H2O |
| 24 | mms6 B9 | 5 PCR sample + 2 Dye + 3 H2O |
| 25 | mms6 F10 | 5 PCR sample + 2 Dye + 3 H2O |
| 26 | rbs-xylE I1 | 5 PCR sample + 2 Dye + 3 H2O |
| 27 | rbs-xylE I2 | 5 PCR sample + 2 Dye + 3 H2O |
| 28 | xylE D7 | 5 PCR sample + 2 Dye + 3 H2O |
| 29 | xylE D8 | 5 PCR sample + 2 Dye + 3 H2O |
| 30 | pLacI-sRBS-lum-dt I8 | 5 PCR sample + 2 Dye + 3 H2O |
| 31 | pLacI-sRBS-lum-dt I9 | 5 PCR sample + 2 Dye + 3 H2O |
| 32 | sRBS-lum-dt A1 | 1 PCR sample + 2 Dye + 9 H2O |
| 33 | sRBS-lum-dt A2 | 1 PCR sample + 2 Dye + 9 H2O |
| 34 | sRBS-lum-dt B7 | 1 PCR sample + 2 Dye + 9 H2O |
| 35 | sRBS-lum-dt B8 | 1 PCR sample + 2 Dye + 9 H2O |
| 36 | sRBS-lum-dt G2 | 1 PCR sample + 2 Dye + 9 H2O |
| 37 | sRBS-lum-dt G3 | 1 PCR sample + 2 Dye + 9 H2O |
| 38 | sRBS-lum-dt A9 | 1 PCR sample + 2 Dye + 9 H2O |
| 39 | sRBS-lum-dt D1 | 0.5 ladder + 2 dye + 9.5 Milli-Q H2O |
| 40 | pSB1A3 control | 5 PCR sample + 2 Dye + 3 H2O |
| 41 | pSB1A3 | 5 PCR sample + 2 Dye + 3 H2O |
| 42 | pSB1A3 control | 5 PCR sample + 2 Dye + 3 H2O |
| 43 | pSB1A3 | 5 PCR sample + 2 Dye + 3 H2O |
Ran gel for 45 minutes at 100V and stained in EtBr for 20 minutes and de-stained for 10 minutes.
- all Econe tubes are red
- all Phusion tubes are blue
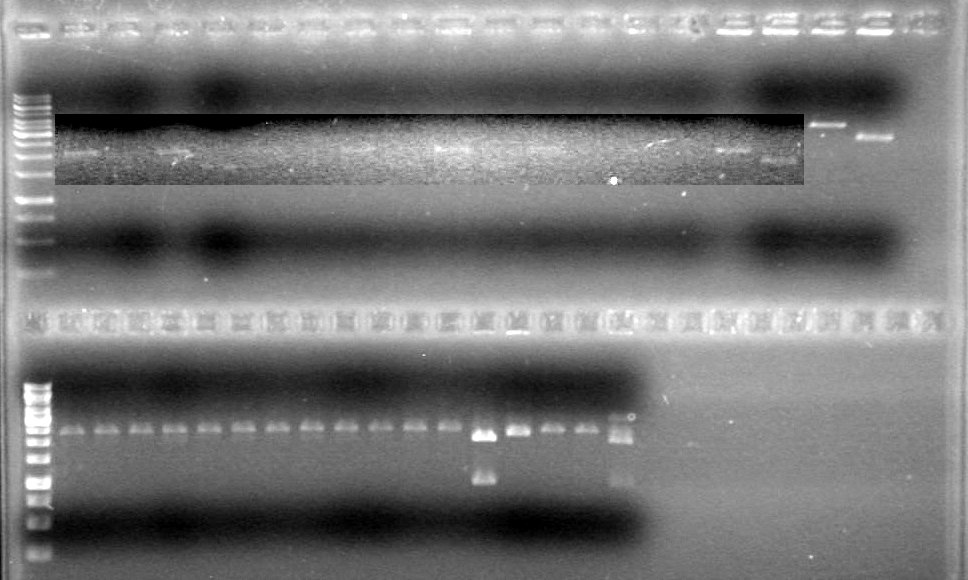
Results:
IMAGE!!!
Objective: Adam put in overnight cultures on June 22. I want to extract the plasmid DNA.
Method:
- Put in overnight cultures at 8:30pm and taken out at 9:30am next morning.
- All cultures were DH5α in LB w/ Amp.
- Cultures were incubated at 37oC.
- Next morning cultures were "minipreped" using boiling lysis miniprep protocol.
Results: Will view plasmid DNA extracted, on a 1% agarose gel (1X TAE) run at 100V for an hour.
| Lane | Sample | Load (µL) |
| 1 | 1kb Ladder | 0.5 Ladder + 2 Dye (6X) + 9.5 Milli-Q H2O |
| 2 | CFP complete 2010 box C8 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 3 | Fusion CEYFP 2007 box H5 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 4 | NEYFP 2007 box J6 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 5 | Fusion CEYFP 2007 box J5 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 6 | CFP complete 2010 box A10 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 7 | pSB-NEYFP 2010 box B6 (C1?) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 8 | Fusion CEYFP 2007 box I5 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 9 | xylE 2007 box B9 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 10 | CEYFP 2007 box G6 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 11 | CFP complete 2010 box A6 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 12 | pSB-CEYFP 2010 box C5 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 13 | pSB-NEYFP 2010 box B6 (C1?) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 14 | pSB-CEYFP 2010 BOX C3 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 15 | xylE 2007 box C4 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 16 | CEYFP 2007 box H6 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 17 | NEYFP 2007 box J6 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
Picture to come!!!!!!!!!
June 24/2010
(in lab: JV, HS, AV)
Objective:: To run a PCR of biobrick parts from out working plasmid box.
Method:
| Master Mix | Volume/tube (µL) | Total Volume (µL) |
| MilliQ H2O | 12.8 | 448 |
| DNA | 1 | ------- |
| 5X Phusion Buffer | 4 | 140 |
| 10µM dNTP's | 1 | 35 |
| Forward Primer | 0.5 | 17.5 |
| Reverse Primer | 0.5 | 17.5 |
| Polymerase | 0.2 | 7 |
Ran for 25 cycles on lig25 setting.
Objective:: Restriction Digest, Run on agarose gel, and ligate PCR products from June 23/10.
Method:
Six tubes for restriction digest:
| Part 1 | Part 2 |
| Lane 8 (I2) | Lane 4 (C1) |
| Lane 19 (A9) | Lane 8 (I2) |
| Lane 19 (A9) | Lane 8 (I2) |
Restriction Digest Reaction Mixture:
| Ingredient | Volume (µL) |
| Plasmid DNA | 2 |
| Enzyme | 0.5 |
| Buffer | 2 |
| Milli-Q H2O | 15.5 |
Incubate at 37oC for 1 hour. Then heat kill the enzymes at 80oC for 20 minutes.
2% Agarose gel electrophoresis:
| Lane | Sample | Load (µL) |
| 1 | 50bp Ladder | 1 Ladder + 2 Dye (6X) + 5 Milli-Q H2O |
| 2 | Fus-RBS-xylE (I2); Lane 8 | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 3 | Fus-RBS-xylE (I2) restricted; Lane 8 | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 4 | Phu-dT (C1); Lane 4 | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 5 | Phu-dT (C1) restricted; Lane 4 | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 6 | Phu-pLacI (A9); Lane 19(1) | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 7 | Phu-pLacI (A9) restricted; Lane 19(1) | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 8 | Phu-pLacI (A9); Lane 19(2) | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 9 | Phu-pLacI (A9) restricted; Lane 19(2) | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 10 | Fus-RBS-xylE (I2); Lane 8(1) | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 11 | Fus-RBS-xylE (I2) restricted; Lane 8(1) | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 12 | Fus-RBS-xylE (I2); Lane 8(2) | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
| 13 | Fus-RBS-xylE (I2) restricted; Lane 8(2) | 5 PCR sample + 2 Dye (6X) + 5 Milli-Q H2O |
Ran gel at 100V for 60 min and stained in EtBr for 15 min.
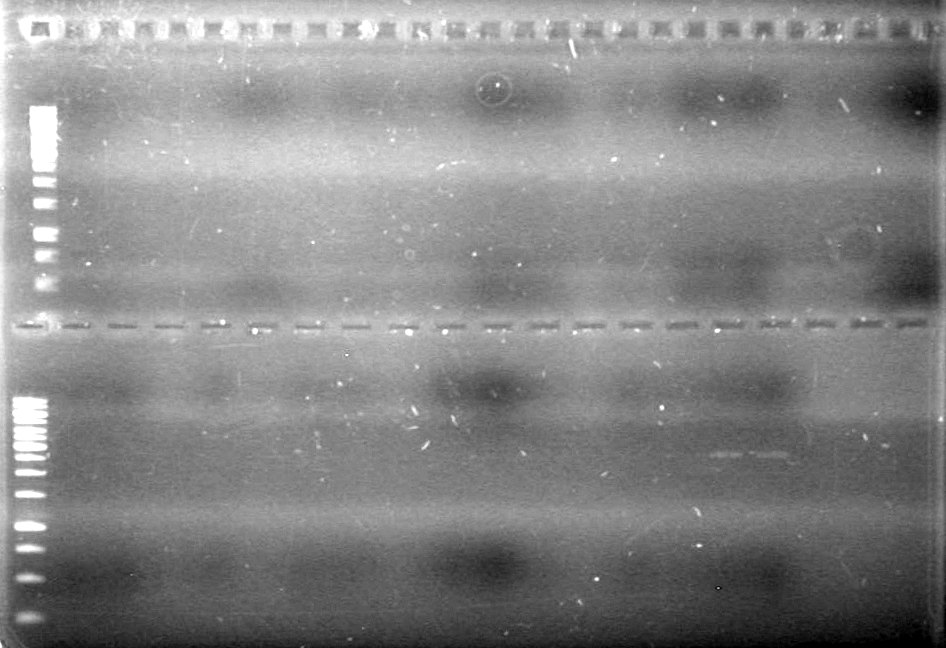
Picture to come!!!!!!!!!
Objective:: Run 1% agarose gel of PCR samples from June 24/10.
| Lane | Sample | Load (µL) |
| 1 | A4-pBAD | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 2 | A4-pBAD | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 3 | A6-SRBS | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 4 | A7-SRBS | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 5 | A8-CFP Complete | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 6 | A10-SRBS | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 7 | B1-EYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 8 | B2-N-term tag | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 9 | B4-pSB-NEYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 10 | B5-pSB-NEYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 11 | B6-CFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 12 | B10-pBAD-TetR | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 13 | D3 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 14 | D4-C-term tag | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 15 | D5-C-term tag | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 16 | D6-PLacI | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 17 | E2-NEYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 18 | E6-CEYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 19 | E7-CEYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 20 | 1kb ladder | 0.5 ladder + 2 Dye (6X) + 9.5 Milli-Q H2O |
| 21 | E8-EYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 22 | E9-EYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 23 | E10-ECFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 24 | F1-ECFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 25 | F2-ECFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 26 | F3-ECFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 27 | F4-pBAD-TetR | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 28 | F5-pBAD-TetR | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 29 | G1-EYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 30 | G4-pSB-CEYFP | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 31 | G6-pBAD(1) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 32 | G7-pBAD(2) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 33 | G8-N-term tag | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 33 | G9-Lumazine | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 33 | 1kb ladder | 0.5 ladder + 2 Dye (6X) + 9.5 Milli-Q H2O |
Results: 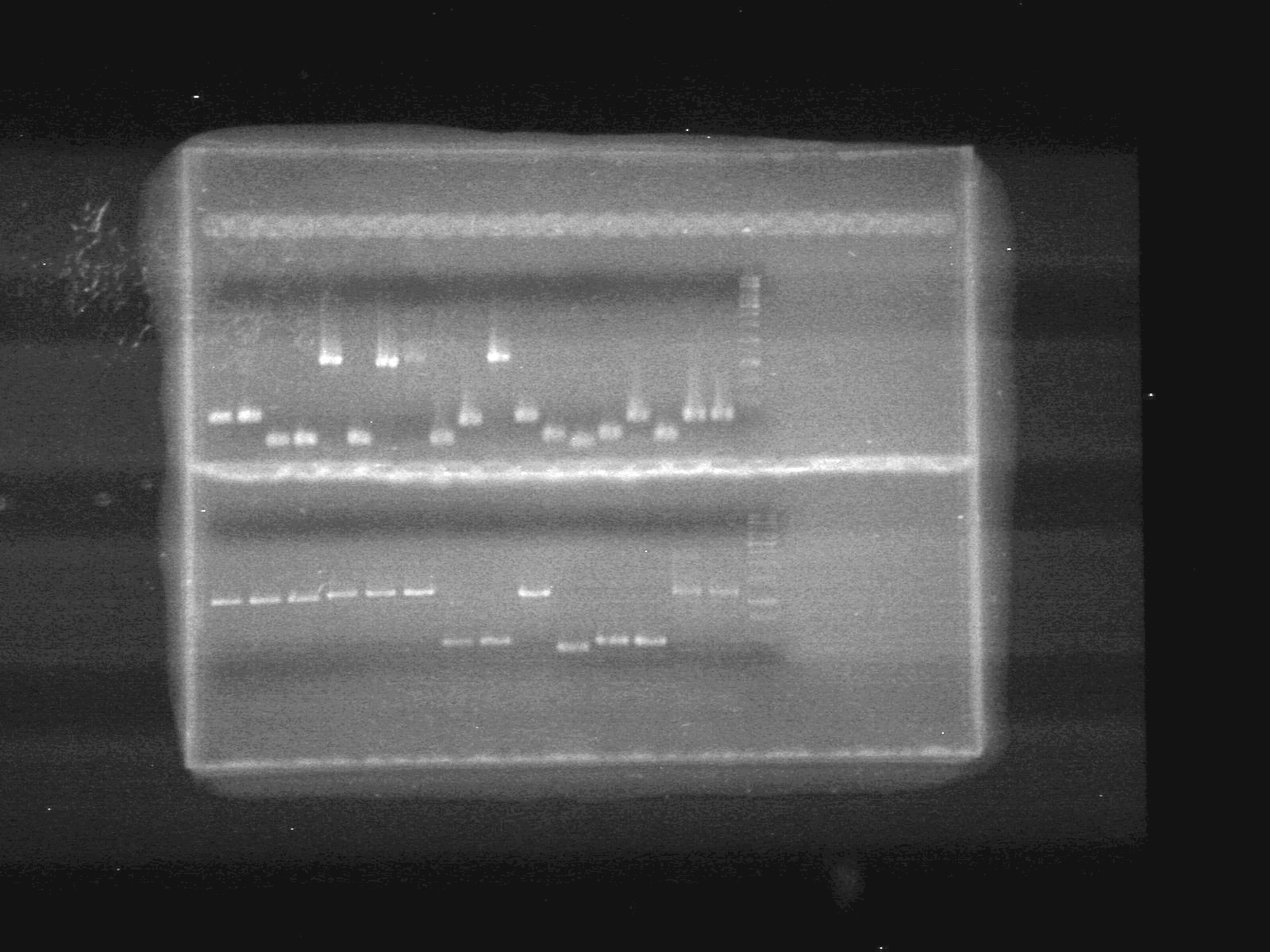
June 28/2010
(in lab: JV, HS, AV, HB)
Objective:: To restrict, run 1% agarose gel, and ligate all mms6, lumazine, xylE into pET28(a) for overexpression tests.
Method:
1) Restrict all parts and pET28(a) with NotI
2) Run on agarose gel (1%)
3) Ligate parts into pET28(a)
Parts:
- from working plasmid box 2010:
- A3-Lumazine
- B9-Mms6
- D7-xylE
- D8-xylE
- I10-Mms6
- from pink tray:
- B9-xylE
- C4-xylE
- G9-Lumazine
- (2X) pET28(a)
Restrictions:
- 2µL DNA
- 2µL Orange Buffer
- 0.25µL Enzyme (NotI)
- 15.75µL Milli-Q H2O
Protocol:
- 1) Incubate for 1 hour at 37oC
- 2) Heat kill for 20 minutes at 80oC (heat block)
Agarose Gel:
| Lane | Sample | Load (µL) |
| 1 | 1kb ladder | 0.5 ladder + 2 Dye (6X) + 9.5 Milli-Q H2O |
| 2 | Lumazine A3 RD | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 3 | Lumazine A3 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 4 | Mms6 I10 RD | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 5 | Mms6 I10 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 6 | Mms6 B9 RD | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 7 | Mms6 B9 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 8 | xylE D8 RD | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 9 | xylE D8 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 10 | xylE D7 RD | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 11 | xylE D7 | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 12 | Lumazine G9 RD (p) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 13 | Lumazine G9 (p) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 14 | xylE B9 RD (p) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 15 | xylE B9 (p) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 16 | xylE C4 RD (p) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 17 | xylE C4 (p) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 18 | pET28(a) RD | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 19 | pET28(a) RD | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
| 20 | pET28(a) | 5 DNA + 2 Dye (6X) + 5 Milli-Q H2O |
Ran gel at 100V for 90 minutes
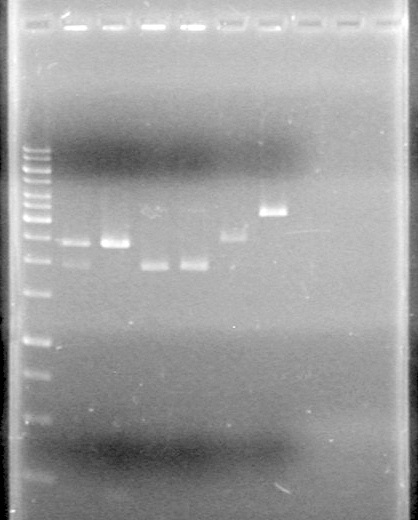
Results:pET28(a) bands are all smeared, and are not useful from inserting parts.
Picture to come!!!!!!!!!
June 30/2010
(in lab: JV)
Objective:: Isolate plasmid DNA from DH5α
Method:Used Kothe Maxiprep protocol.
Cell Pellet Weights:
- xylE = 1.15g
- Mms6 = 1.13g
- pET28(a) = 0.67g
- lumazine synthase = 1.1g
Results:
 "
"