Team:SDU-Denmark/labnotes6
From 2010.igem.org
Lab notes (16/9 - 22/9)
Contents |
Group: Retinal
ON of Top 10 E. coli cells with POT2 with NinaB
Date: 16/8
Done by: Marie & Tommy
Methods:
protocos:
Notes:
100 mL ON culture was made, cells were grown at 37 C in LB media with Chloramphenicol.
ON of Top 10 E. coli cells with POT2 with NinaB
Date: 17/8
Done by: Marie & Tommy
Methods: Miniprep, Restriction digest, gel
Protocols: MP1.2, RD1.1.
Notes:
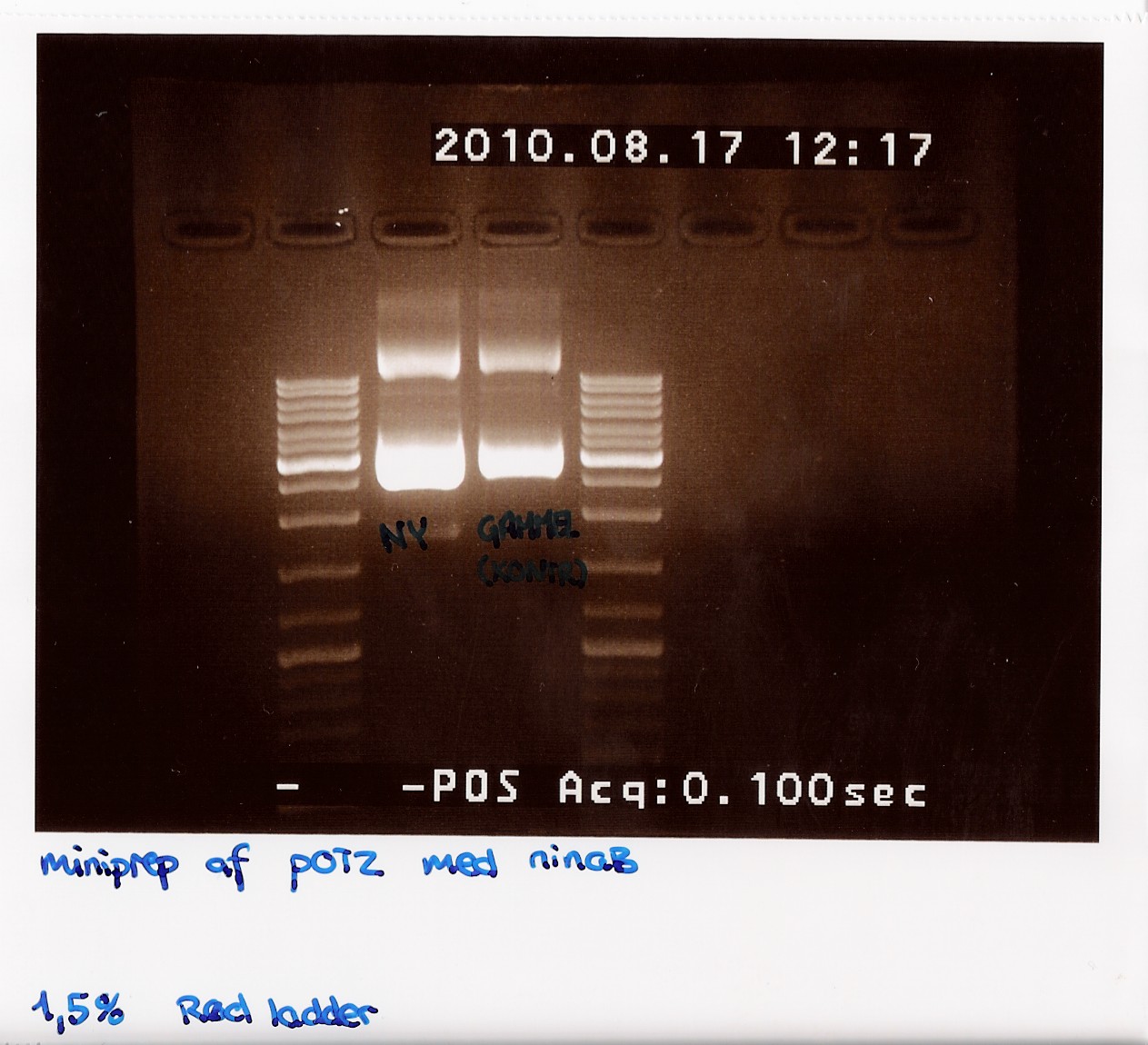
50 mL of elution buffer was used. DNA concentrations after pooling were measured on NanoDrop to 206,2 ng/microL.
gel was run on produckt, showing recent miniprep compared to old miniprep produckt, showing uneven lengths. subsequent was performed.
Restriction digest on miniprep produckt (w. EcoRI)
Date: 17/8
Done by: Marie & Tommy
Methods: Restriction digest, gel
Protocols: MP1.2, gel.
Notes:
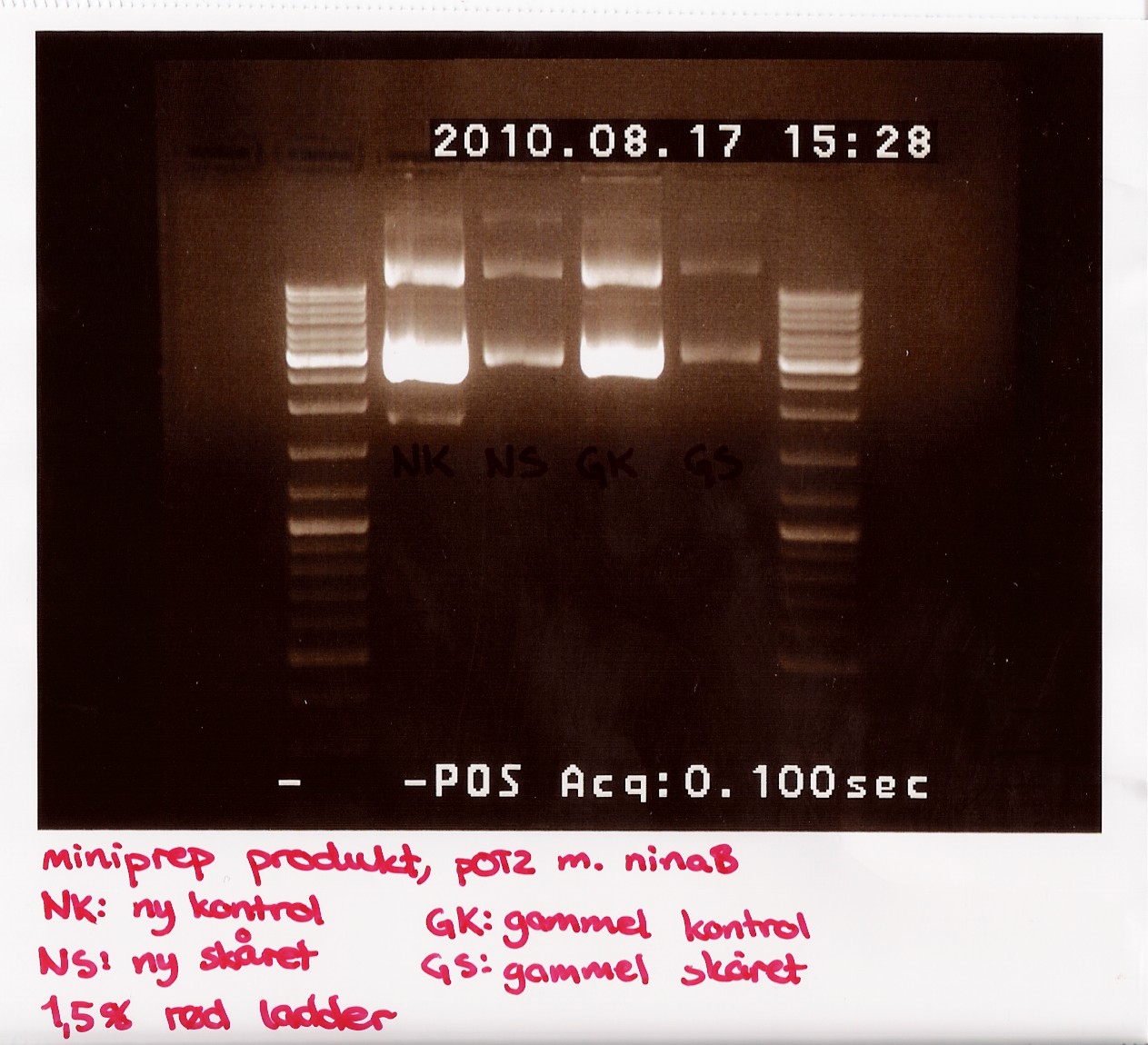
Due to the lack of old sample, restriction digest was performed using only 3,5 microL of miniprep produckt. 1,5 microL of H2O was added insted. EcoRI was used
The digest mix was incubated for 10 min at 37 C. A gel was run showing uncut new, cut new, uncut old and cut old miniprep product.
PCR on POT2 with NinaB (New Primers)
Start date: 20/8
Methods: PCR, Gel
Protocols: CP.1.1
PCR on POT2 with NinaB (New Primers)
Date: 20/8
Done by: Marie & Tommy
Methods: ON
protocos:CP1.1
Notes:
The new primers only contain the innermost restriction sites.
Meltting temp.: FWD: 68,1 C REV: 65,6 C
To get the optimal PCR temperatures a gradient PCR were run programed as:
| PCR | Temp. (C) | Time (min) |
| Start | 95 | 2 |
| Denaturing | 95 | 1 |
| Anneling | Gradient | 1 |
| Elongation | 72 | 4 |
| End | 72 | 5 |
| Hold | 4 | indef. |
The temperatur gradient were run from 60 to 70 C and the samples were run at these temperatures:
| Sample | Colunm | Temp. C |
| 1 | 1 | 59,9 |
| 2 | 3 | 60,7 |
| 3 | 5 | 62,7 |
| 4 | 7 | 65,4 |
| 5 | 9 | 67,9 |
| 6 | 11 | 69,6 |
After the PRC the product were run on a gel
The gel picture shows that alle of the temperatures gave results.
 "
"