Team:SDU-Denmark/safety-d
From 2010.igem.org
Useful ideas for Improving Safety
Watermarking
To increase public safety we propose to introduce a water-marking standard.
Following the example of J. Craig Venter, who, in may 2010, created the first watermark in a bacteria, containing several readable messages, we propose to create a watermarking standard to increase the safety of the environment, as well as the safety of the community at large. <vr>
But why should we consider creating and using a watermarking standard for work in synthetic biology? Let us consider the following example:
A company has created a synthetic organism capable of absorbing harmful substances from the ground. The government, eager to help clean polluted ground, releases the bacteria into the wild, confident that the bacteria will not pose any threat to the environment. These bacteria do not, however, react as planned. Instead of absorbing only the harmful substances from the ground, they transfer the substances to other organisms, or simply run amok spreading themselves in an uncontrolled manner, causing harm instead of good. Let us imagine that this happens close to the nation's border. The government in the neighboring state finds itself with a unexplained biological phenomenon, possibly causing great harm.
Should the rogue bacteria contain engineered, watermarked parts, it would be a small matter to have the parts sequenced, thus the government would be able to easily access all the relevant information on the rogue bacteria, contact the manufacturers and would know how to stop it.
The watermark
We believe that a watermark should contain the following:
I. A 12 nucleotide ‘license’
We have set the following criteria for a good watermark:
I. It should contain the creating team’s ‘license’
II. It should not interfere with the other functions of the part
III. It should be persistent in the plasmid, i.e. not be removed from the plasmid due to natural evolution
IV. It should be easy to find, easy to read and easy to insert by the developing team
This 12 nucleotide license would then refer to a database where the following information should be available:
I. It should contain information on the creating team
II. The name of the part
III. A description of the part
IV. A risk-assessment of the part by the creating team
V. Information on how to neutralize organism, and, if available, kill-code
Placing the watermark
The initial thought was to insert the watermark into the genome of the bacteria, so as to increase the stability of the watermark in the bacteria. However, as all parts inserted into the bacteria are placed in plasmids, it would make no sense to insert the watermark into the genome. The bacteria could transfer their plasmids to other bacteria, and retain their watermark, and the watermark would have become useless, persisting in bacteria that have no modified material at all.
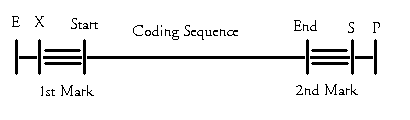
Also, the thought was to mark the bacteria, and not the parts. Thus we could have a single watermark to cover the entire modified organism. However, this presents us with many of the above problems. Should anyone encounter a rogue bacteria which has lost some of its plasmids, the watermark would be useless. It would only cover the entire modified system, and should some, or all bacteria have discarded the plasmids, the watermark would be useless. We therefore propose to mark every plasmid, before and after the coding sequence. The watermark will therefore be split into two separate components. The team creates a watermark for every part, and inserts it into the plasmid, before and after the coding sequence.
We chose to divide the code into two separate parts for two reasons. Firstly, being split into two equally long parts at each end of the coding sequence ensures symmetry , which again should help make insertion of a watermark easy, as this will reduce the complexity of the primers we need to design to insert them. Secondly this ensures that identification is done easily, since the combination of nucleotides of the sequence from E to X and from S to P is known. Thus we effectively increase the sequence we search for from 6 to 26 nucleotides. Also this means that the risk of a naturally occurring combination of nucleotides, identical to the synthetically created one decreases.
Alternative placement
We also thought about the possibility of placing the watermark between the cutting sites of E and X, and S and P respectively. Since this area is a spacing area, it might be a good spot to insert the nucleotide combination we want. Difficulties in this might cause disturbance. Another idea was to expand the restriction sites E-X and S-P, but we are uncertain whether this might disturb the functionality of the part.
Size and design
Placing the watermark after the restriction sites, it should be relatively small, as to not interfere with the functionality of the part into which it is inserted. We propose that the watermark should be 12 nucleotides, divided in two groups of six which will allow for 16.777.216 combinations of A, C, G and T in total.
The watermark must not contain any restriction-enzymes or stop codons. If the watermark accidentally contained a restriction-enzyme or a stop codon, the watermark would interfere with the function of the part into which it is inserted, and would likely render the part useless. This severely restricts the number of combinations we can use.
Ideally we would have liked to use more nucleotides, as we would have been able to generate more combinations. But the watermark should be as small as possible as not to interfere with the functions (i.e. cause a frame shift) of the part and not make the design of primers unduly complicated. We could make relatively long watermarks to satisfy our need for a very large number of possible combinations, but it would make the design of the necessary primers extremely complicated, and would go against our goal of making the insertion of watermarks as small and easy a procedure as possible. We believe that the best compromise between the amount of combinations and the ease of insertion would be at around 12 nucleotides.
License
Every team and lab/company which works with synthetic biology will be assigned a specific “license plate”, unique to them and being their ‘ID’ on parts-registry. An extension to parts registry’s search function should then be added. If its database contains information on each “license”, one could easily find information on a foreign organism, in case they found it in the wild (that is, anywhere else than the lab), and had it sequenced afterwards. Upon entering the parts-registry, one should then be able to enter the license and gain access to information on the parts that the team / lab / company has created. Anyone should be able to enter the license code into the parts-registry and gain access to all information on the creating team, the parts created by the team, and of course some contact information, in order to seek advice.
Procedure
Retrieval of the watermark should be easy, in the event of a rogue bacteria spreading havoc in the environment. Standardized placement of the watermark within the part should make it possible to easily retrieving the watermark through sequencing. Thus, if a rogue bacteria is discovered, it should be a simple matter to sequence the part, and obtain the license. Then it is just a matter of consulting the parts-registry to access all available information on the part and on how to contact the creating team. Getting a bacteria sequenced is a technology which is easy to access, and a lot of companies worldwide offers sequencing of bacteria, in addition it is a relative speedy process, which further makes the identification of a malign bacteria, and its properties relatively easy.
Questions for assessing parts
Here is a list of questions that we believe the team should ask itself and answer, to its best ability when considering what information is relevant for the safety and security of their project.
General use
What type of lab should work with this BioBrick be done in?
Is any special care needed when working with this BioBrick in addition to ordinary laboratory work-safety protocols?
Potential pathogenicity
Does this BioBrick produce any product that is toxic to plants or animals?
Could a host with this BioBrick produce sufficient amounts of an otherwise non-toxic product to toxic levels?
Is the gene that the BioBrick codes for associated with disease processes or infection in humans?
Does the gene that the BioBrick codes for have homologs in known pathogenic bacteria? What are these genes’ functions?
Does this BioBrick produce any product that can regulate the immune system in animals or humans?
Environmental impact
Does this BioBrick produce anything that plays an significant role in environmental processes?
Does this BioBricks increase it’s hosts ability to outmatch naturally occurring bacteria? Does it disrupt natural occurring symbiosis?
Does this BioBrick increase it’s host’s ability to replicate?
Does this BioBrick increase it’s host’s ability to spread?
Does this BioBrick increase it’s host’s ability to survive outside laboratory conditions?
Does this BioBrick create something that might be toxic to it’s environment?
Possible malign use
Can host’s with this BioBrick survive storage conditions (in pressured conditions, under alternative temperatures and in large containers?)
Can your chosen host be arosoled?
Does this BioBrick increase the host’s ability to vaporize?
Does this BioBrick increase the host’s ability to create spores?
Does this BioBrick produce any product that can regulate the immune system in animals or humans?
Is this BioBrick in anyway pathogenic in animals or humans or towards plants?
Construct notes
What is the origin of the genetic material used? What does the the genetic materiale do in this origin? Is there uncertainty about the genetical materials function?
What modifications were done on the genetic materiale before insertion? If anything was modified, what function do you hope to achieve?
What vector did you use? Which antibiotic resistance were involved? Which protocol was used to insert the vector?
What is the stability of the insert with respect to genetic traits?
How easily can the insert transfer to other bacteria or lifeforms?
Where there safer alternatives to achieve this function? Where there safer alternatives to the host organism and vector used?
Is your construct watermarked?
The above list of question is more or less derived from a more throughout list of questions relevant to assessing the safety and securty of the parts, see appendix II.
Further Information
Risk-assessment for organism
Should the part, or a number of parts, be inserted into an organism the team should perform a risk-assessment and make it available on the parts-registry. In some countries, it is mandatory to submit a risk-assessment prior to engaging in a project involving synthetic biology, so we believe that any risk-assessments should be made public through parts-registry.
Inclusion of copyright information?
We do not believe in any form of copyright prohibition. We believe in an open-source approach to the field of synthetic biology, as in iGEM. Any copy-right prohibitions would only stall the progress in this most vital field of science. We believe that any and all information on created parts, and experience with these parts in particular organisms, should be shared freely.
Information on how to neutralize bacteria
This clause is intended as security measure. Should the bacteria be released into the environment, the parts-registry site should contain information on how to neutralize the bacteria. If the bacteria has an kill-code inserted, the site should describe how to enact the self-destruct mechanism.
Anticipated problems
Code deterioration
The code will deteriorate over time due to mutation. This could prove to be a serious problem should watermarking become an integrated part of synthetic biology. Should the genetic watermark deteriorate to the point where one is no longer able to read it, it would not constitute any kind of safety measure, being able to tell that the part was likely to have been made artificially being the only thing we would be able to tell. Knowing that data will deteriorate, it may be impossible to determine whether the watermark found in rogue bacteria is authentic or a degenerate. The deterioration is however slow and arbitrary. Our code is so small, that the change that any nucleotide associated with the watermark is going to mutate is very limited. The chances of finding the authentic code intact should be very good.
The open source approach
Sharing all this information on creating new synthetic parts that can be inserted into living organisms, also means that people with harmful agendas have access to this knowledge. This naturally means that wrongful actions are possible by use of synthetic biology, but we do not think that this should stand in the way of all the possibilities synthetic biology holds. Also, the more we know about how to create synthetic DNA strands, the better we are equipped if any harmful incident should occur. As of now, it might also just be easier to drop a bomb or send out letters of anthrax. Should a bacteria be used for a malign purpose it would be quite easy to insert a false watermark to blame others. So we need to keep in mind that a plot could be made against someone. However, if anyone was interested in harming as many as possible, this person probably wouldn’t care about watermarking at all. This also means that we cannot expect watermarking to play a part in any legal case.
 "
"