Team:SDU-Denmark/labnotes10
From 2010.igem.org
Lab notes (9/13 - 9/19)
Photosensor group
Control restriction digest of PS+B0015 in pSB1AK3 sample Q, miniprep
Date: 09/14
Done by: LC
Methods: Restriction digest
Protocols: RD1.1[1]
Notes:
The afore mentioned miniprep of PS+B0015 in pSB1AK3 (Q) was cut with EcoRI and PstI to check if biobrick pre- and suffix are present and the insert is the right size.
Mix for 1 RD reaction:
12 ul H2O
1 ul PstI
1 ul EcoRI
2 ul Fast digest green buffer
5 ul PCR product
Uncut PCR product was loaded on the gel as a control.
Results: The RD resulted in three bands, uncut plasmid, plasmid and insert. All of those were the expected lengths, so the sample should be OK.

Taq PCR of miniprep of pSB1C3 w. PS col. B and pSB1AK3 w. PS + double termiantor col. C
Date: 9/15 2010
Done By: Maria and Lc
Protocol:CP1.3
Notes:
To ensure that the plasminds sent to sequencing contains the correct inserts, af controle PCR of the minipreps using the PS forward and reverse primer is carried out.
Premix x 3
7.5uL Taq buffer
3uL MgCl2
3uL PS FW
3uL PS RV
1.5uL dNTP
52.2uL H2O
1uL of template is distrubuted into each PCR tube.
PCR program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
2:30 min |
| Goto2 |
rep |
29x |
| End |
72 C |
5 min |
| Hold |
4 C |
The PCR product was loaded onto a 1.5% agarose gel. Gene ruler DNA ladder mix was used as marker.
Results:
Analysis:
2 bands at app. 3000bp and 4500bp respectively, corresponds to the template sizes, indicating that too much template has been used.
--Tipi 06:44, 28 September 2010 (UTC)
Insertion of PS + double terminator in pSB3T5 with J13002
Date: 9/13 - 9/16 2010
Done By: Maria and Lc
Protocol: CP1.1DE1.3RD1.1LG1.2CC1.1TR1.1CP1.3
Pfu PCR amplification of PS + double terminator (no.1)
Date: 9/13 2010
Done By: Maria and Lc
Protocol:CP1.1
Notes:
6 PCR reactions are prepared. 3.5uL Miniprep of PS+ double terminator in pSB1AK3 are diluted in 3.5uL H2O to reach a 2x dilution, and are used as template. PCR tubes are marked PS+B0015.1 A-F.
Premix x6:
| pfu buffer + MgSO4 | 35uL |
| dNTP mix | 10.5uL |
| VF2 primer | 10.5uL |
| VR primer | 10.5uL |
| H20 | 273uL |
| pfu polymerase | 3uL |
| PS + Double terminator in pSB1AK3 (2x diluted) | 7uL |
PCR program:
| start | 95C | 3min |
| denaturating | 95C | 2min |
| annealing | 55C | 30s |
| elongation | 72C | 4min30s |
| go to | 2 | rep.29x |
| end | 72C | 5min |
| hold | 4C |
All of the PCR product was loaded onto a 1.5 agarose extraction gel. Gene ruler DNA ladder mix was used ad marker.
Results:
Analysis:
Bands were observed at app. 2500bp and bands were extracted by gel extraction
Gel extraction of PS + double terminator
Date: 9/13 2010
Done By: Maria and Lc
Protocol:DE1.3
Notes:
DNA was extracted from gel according to protocol and each sample was diluted in 20uL.
Analysis:
samples were pooled and used for restriction digest.
Restriction digest of PS + double terminator, and PSB3T5 no. 1
Date: 9/13 2010
Done By: Maria and Lc
Protocol:RD1.1DE1.3
Notes:
Restriction mixture pSB3T5:
| H2O | 12uL |
| FD green buffer | 2uL |
| SpeI | 1uL |
| PstI | 1uL |
| pSB3T5 | 5uL |
Restriction mixture PS + double terminator:
| H2O | 38uL |
| FD green buffer | 8uL |
| XbaI | 4uL |
| PstI | 4uL |
| PS + double terminator | 30uL |
Digested samples were loaded onto a 1.5% agarose extraction gel. Uncut PS + double terminator and pSB3T5 were used as controles. Gene ruler DNA ladder mix was used as marker.
Loading scheme:
| Lane | sample |
| 1 | PS + double terminator |
| 2 | uncut PS + double terminator |
| 3 | pSB3T5 |
| 4 | uncut pSB3T5 |
| 5 | marker |
DNA was extracted from gel, however undiluted washing buffer was used and the DNA in the samples was destroyed.
Pfu PCR amplification of PS + double terminator (no.2)
Date: 9/13 2010
Done By: Maria and Lc
Protocol:CP1.1
Notes:
6 PCR reactions are prepared. 3.5uL Miniprep of PS+ double terminator in pSB1AK3 are diluted in 3.5uL H2O to reach a 2x dilution, and are used as template. PCR tubes are marked PS+B0015.1 A-F.
Premix x6:
| pfu buffer + MgSO4 | 35uL |
| dNTP mix | 10.5uL |
| VF2 primer | 10.5uL |
| VR primer | 10.5uL |
| H20 | 273uL |
| pfu polymerase | 3uL |
| PS + Double terminator in pSB1AK3 (2x diluted) | 7uL |
PCR program:
| start | 95C | 3min |
| denaturating | 95C | 2min |
| annealing | 55C | 30s |
| elongation | 72C | 4min30s |
| go to | 2 | rep.29x |
| end | 72C | 5min |
| hold | 4C |
All of the PCR product was loaded onto a 1.5 agarose extraction gel. Gene ruler DNA ladder mix was used ad marker.
Results:
Analysis:
Bands were observed at app. 2500bp and bands were extracted by gel extraction
Gel extraction of PS + double terminator
Date: 9/14 2010
Done By: Maria and Lc
Protocol:DE1.3
Notes:
DNA was extracted from gel according to protocol and each sample was diluted in 20uL.
Analysis:
samples were pooled and used for restriction digest.
Restriction digest of PS + double terminator, and PSB3T5 (no.2)
Date: 9/14 2010
Done By: Maria and Lc
Protocol:RD1.1DE1.3
Notes:
Restriction mixture pSB3T5:
| H2O | 14uL |
| FD green buffer | 4uL |
| SpeI | 2uL |
| PstI | 2uL |
| pSB3T5 | 20uL |
Restriction mixture PS + double terminator:
| H2O | 38uL |
| FD green buffer | 8uL |
| XbaI | 4uL |
| PstI | 4uL |
| PS + double terminator | 30uL |
Digested samples were loaded onto a 1.5% agarose extraction gel. Uncut PS + double terminator and pSB3T5 were used as controles. Gene ruler DNA ladder mix was used as marker.
Loading scheme:
| Lane | sample |
| 1 | Marker |
| 2 | uncut PS + double terminator |
| 3 | PS + double terminator |
| 4 | uncut pSB3T5 |
| 5 | pSB3T5 |
DNA was extracted from gel, according to protocol. Samples were diluted in 20uL H2O. 2. dilution of PS + doubletermiantor was diluted in 10uL H2O
Results:
DNA conc:
| sample | conc. (ng/uL) |
| PS + double terminator (1. dilution) | 4.8 |
| PS + double terminator (2. dilution) | 2.3 |
| pSB3T5 | 26.4 |
Analysis:
the purified DNA was used for ligation.
Ligation of PS + double terminator and pSB3T5
Date: 9/14 2010
Done By: Maria and Lc
Protocol:LG1.2
Notes:
Three ligation mixtures was prepared. vector concentrations of 25ng/uL was used for each mixture. Appropiate amount of insert was added to reac vector:insert ratios of 1:1, 1:2 and 1:3 respectively.
Ligation mixtures (PS + double terminator in pSB3T5):
| L1 | L2 | L3 | |
| T4 ligase buffer | 2uL | 2uL | 2uL |
| T4 ligase | 1uL | 1uL | 1uL |
| pSB3T5 | 1uL | 1uL | 1uL |
| PS + double terminator | 8uL (2. dilution) | 7uL | 11uL |
| H20 | 8uL | 9uL | 5uL |
The samples were incubated at 17C ON at used for transformation
Transfomation of ligated plasmid in Top 10 E.coli
Date: 9/15 2010
Done By: Maria and Lc
Protocol:CC1.1TR1.1
Notes:
The compotent cells and transformation was carried out according to protocol.
Results:
the controle plates were okay, and colonies of different sizes was observed on plates of cells transformed with ligations.
Analysis:
The transformation was successfull and colonies was selected and used in coloni PCR.
--Tipi 13:41, 26 September 2010 (UTC)
Transfomation of pKJ606 in Mg 1655 E.coli
Date: 9/15 2010
Done By: Maria and Lc
Protocol:CC1.1TR1.1
Notes:
pKJ606 plasmid was transformed into Mg1655, to use for characterization.
The compotent cells and transformation was carried out according to protocol.
Results:
the controle plates were okay, and large colonies was observed on the plates.
Analysis:
The transformation was successfull and colonies was selected and used in coloni PCR.
--Tipi 05:48, 28 September 2010 (UTC)
Colony PCR of transformation of pKJ606 in Mg1655 and pSB3T5 w. PS + double terminator in top10 (no.1)
Date: 09/16
Done by: LC & Maria
Methods: Colony PCR
Protocols: CP1.3[2]
Notes:
for the pKJ606 coloni PCR two colonies are selected.
16 colonies of different sizes are selected from the ligation plates. The sizes of the colonies are noted down to test whether this can be used to select colonies containing the correct plasmid
| coloni sizes | large | small |
| colonies | K,B,A,C,D,E,F,I,J,N,O,P | L,G,H,M |
Premix for pKJ606 (x3) :
7.5 µl 10xTAQ Buffer
3 µl MgCl2
3 µl FW PS
3 µl PV PS
1.5 µl dNTP
10.5 µl H2O
1 µl TAQ Polymerase
Premix for pSB3T5 (x17) :
42.5 µl 10xTAQ Buffer
17 µl MgCl2
17 µl VF2
17 µl VR
8.5 µl dNTP
59.5 µl H2O
2 µl TAQ Polymerase
The chosen colonies were lysed in 15 ul of H20.
Both of the coloni PCR's were run using the same program.
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
2:30 min |
| Goto2 |
rep |
29x |
| End |
72 C |
5 min |
| Hold |
4 C |
PCR product was loaded onto a 1.5% agarose gel. Gene ruler DNA ladder mix was used as marker.
Results:
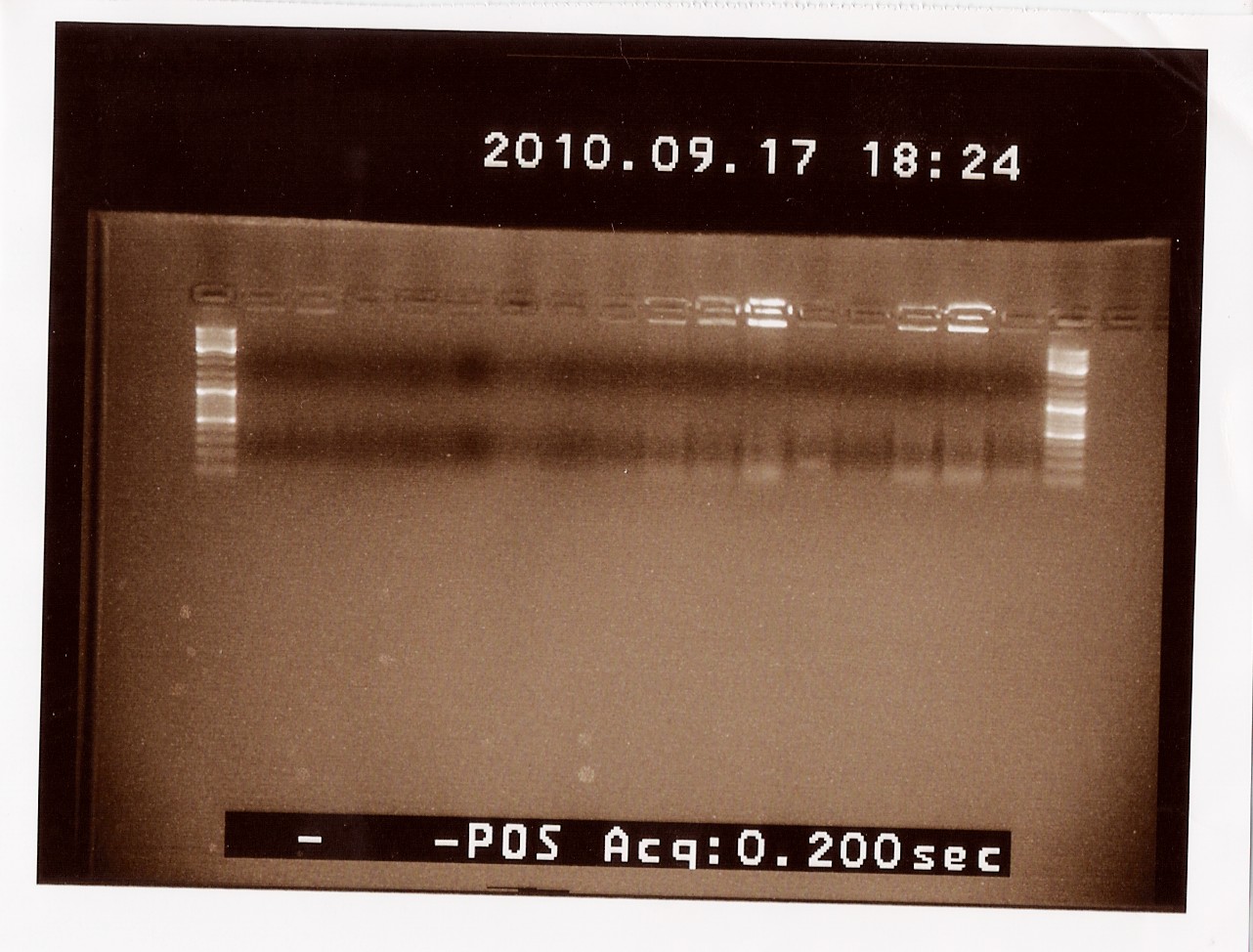
Analysis:
No bands was obeserved on the gel, and the experiment was redone.
--Tipi 06:25, 28 September 2010 (UTC)
Colony PCR of transformation of pKJ606 in Mg1655 and pSB3T5 w. PS + double terminator in top10 (no.2)
Date: 09/16
Done by: LC & Maria
Methods: Colony PCR
Protocols: CP1.3[3]
Notes:
for the pKJ606 coloni PCR two colonies are selected.
16 colonies of different sizes are selected from the ligation plates. The sizes of the colonies are noted down to test whether this can be used to select colonies containing the correct plasmid
| coloni sizes | large | small |
| colonies | O,L,G,A,B | P,N,K,I,H,E,M,J,F,D |
Premix for pKJ606 (x3) :
7.5 µl 10xTAQ Buffer
3 µl MgCl2
3 µl FW PS
3 µl PV PS
1.5 µl dNTP
10.5 µl H2O
1 µl TAQ Polymerase
Premix for pSB3T5 (x17) :
42.5 µl 10xTAQ Buffer
17 µl MgCl2
17 µl VF2
17 µl VR
8.5 µl dNTP
59.5 µl H2O
2 µl TAQ Polymerase
The chosen colonies were lysed in 15 ul of H20.
Both of the coloni PCR's were run using the same program.
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
2:30 min |
| Goto2 |
rep |
29x |
| End |
72 C |
5 min |
| Hold |
4 C |
PCR product was loaded onto a 1.5% agarose gel. Gene ruler DNA ladder mix was used as marker.
Results:
Analysis:
In the lanes containing PCR product of colonies transformed with pKJ606 bands at app. 2000bp are observed, indicating that the colonies contains the right plasmids. No bands was observed in lanes containing PCR product of colonies transformed with pSB3T5, and this experiment needs to be redone.
--Tipi 06:25, 28 September 2010 (UTC)
Insertion of PS + double terminator in pSB3T5 with J13002 (no.2)
Date: 9/16 - 9/19 2010
Done By: Maria and Lc
Protocol: CP1.1DE1.3RD1.1LG1.2CC1.1TR1.1CP1.3
PFU PCR on PS+B0015 with VF2 and VR primers
Date: 9/16 - 9/19 2010
Done By: Maria and LC
Protocol: CP1.3
Notes:
Premix:
30 ul PFU Buffer + MgSO4
9 ul dNTP mix
9 ul VF2
9 ul VR
10 ul template (5 ul of PS in pSB1AK3 diluted in 5 ul H2O)
230 ul H2O
2,5 ul PFU polymerase
Program:
| start | 95C | 3min |
| denaturating | 95C | 2min |
| annealing | 55C | 30s |
| elongation | 72C | 5 min |
| go to | 2 | rep.29x |
| end | 72C | 5min |
| hold | 4C |
Results: The band showed up at the correct length as expected. The remaining product was then extracted from the PCR mixture.
Restriction digest of PS + double terminator and PSB3T5
Date: 9/17 2010
Done By: Maria and Lc
Protocol:RD1.1DE1.3
Notes:
Restriction mixture pSB3T5:
| H2O | 14uL |
| FD green buffer | 4uL |
| SpeI | 2uL |
| PstI | 2uL |
| pSB3T5 | 20uL |
Restriction mixture PS + double terminator:
| H2O | 38uL |
| FD green buffer | 8uL |
| XbaI | 4uL |
| PstI | 4uL |
| PS + double terminator | 30uL |
Samples were digested for 10min. at 37C.
Digested samples were loaded onto a 1.5% agarose extraction gel. Uncut PS + double terminator and pSB3T5 were used as controles. Gene ruler DNA ladder mix was used as marker.
Loading scheme:
| Lane | sample |
| 1 | Marker |
| 2 | pSB3T5 |
| 3 | uncut pSB3T5 |
| 4 | PS + double terminator |
| 5 | uncut PS + double terminator |
DNA was extracted from gel, according to protocol. Samples were diluted in 20uL H2O.
Results:
DNA conc:
| sample | conc. (ng/uL) |
| PS + double terminator | 26.3 |
| pSB3T5 | 25.8 |
Analysis:
the purified DNA was used for ligation.
Ligation of PS + double terminator and pSB3T5
Date: 9/17 2010
Done By: Maria and Lc
Protocol:LG1.2
Notes:
Three ligation mixtures was prepared. 50ng of vector was used for each mixture. Appropiate amount of insert was added to reac vector:insert ratios of 1:1, 1:2 and 1:4 respectively.
Ligation mixtures (PS + double terminator in pSB3T5):
| L1 | L2 | L3 | |
| T4 ligase buffer | 2uL | 2uL | 2uL |
| T4 ligase | 1uL | 1uL | 1uL |
| pSB3T5 | 2uL | 2uL | 2uL |
| PS + double terminator | 1uL (2. dilution) | 2.5uL | 5uL |
| H20 | 14uL | 12.5uL | 10uL |
The samples were incubated at 17C ON at used for transformation
Transfomation of ligated plasmid in Top 10 E.coli
Date: 9/15 2010
Done By: Maria and Lc
Protocol:CC1.1TR1.1
Notes:
The compotent cells and transformation was carried out according to protocol.
Results:
the controle plates were okay, and colonies of different sizes was observed on plates of cells transformed with ligations.
Analysis:
The transformation was successfull and colonies was selected and used in coloni PCR.
--Tipi 11:21, 28 September 2010 (UTC)
Insert clever content here.
 "
"