Team:Washington/Gram Positive/Test
From 2010.igem.org
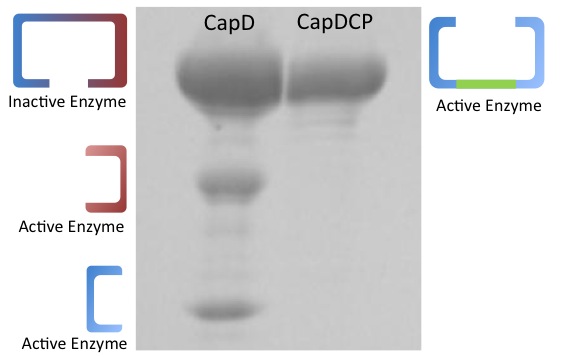
More descriptive titles, verification that your CapD_CP was a monomer ( gel) should go before testing catalytic ability
Contents |
Why CapD_CP?
Easy Quantification
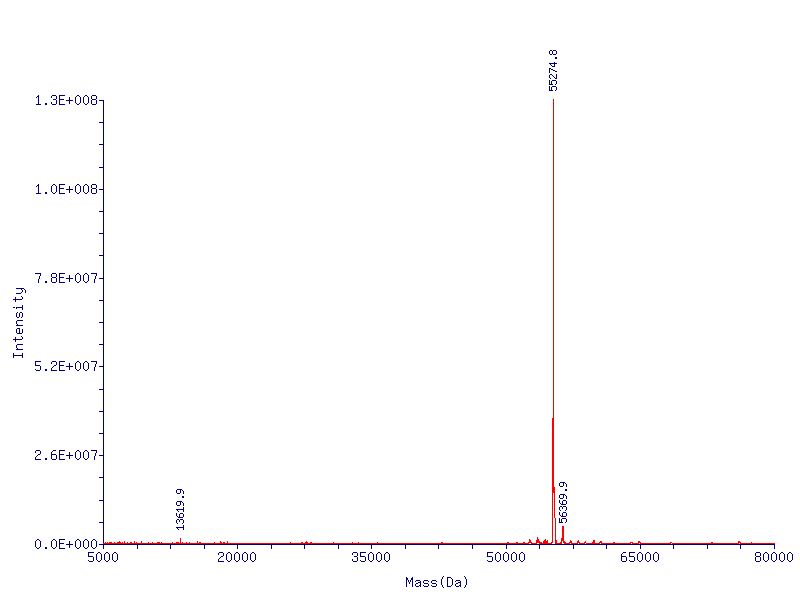
In addition to the fact that CapD_CP is easy to express, it has one more crucial advantage over CapD. When purifying CapD_CP, given our mass spec data, we can assume a massive majority of the purified CapD_CP is functional. CapD, however, is a much more ambiguous case, as illustrated by our protein gel results. The lower two bands of CapD correspond to the two subunits of cleaved CapD, which are active. The upper band may be uncleaved, therefore unfunctional, CapD, or dimeric, active CapD that simply did not denature, or a combination of both. Because of this, its difficult to quantify how much active protein is in a solution of purified CapD, making assaying activity a nightmare. For this reason, we made our active site mutations to CapD_CP.
Unchanged Properties
Test: Enzyme Assay

After we have the CapD_CP mutants, we tested our mutants for their catalytic activity using our fluorescence-based enzyme assay scheme. Fluorescence-based enzyme assay measures the rate at which fluorescence in the testing media is released and the amount fluorescence depends on the rate at which fluorophore-quencher linkage is disrupted. Our substrate PDGA contains a linked fluorophore-quencher component. The faster fluorophore-quencher component is cleaved, the higher the amount of fluorescence is released and thus the greater the enzymatic activity observed.
Results: Data Analysis
Validating CapD_CP Activity
Before we can predict which mutations increase hydrolysis capability, we need to validate that the circularly permuted version of CapD has measurable activity for further assessments. We also hypothesize a threonine residue in the catalytic site of CapD_CP plays an important role in the catalysis reaction and mutating it will eliminate all enzymatic activity. Thus we created two mutants, T2V and T2A, to act as negative controls. The result (figure 1 below) of this assay confirms our hypothesis that CapD_CP has enzymatic activity to the two catalytic knockouts. The relatively flat activity curves of the knockout mutants confirm the hypothesis of the threonine's role in the catalytic site.
Analyzing CapD_CP
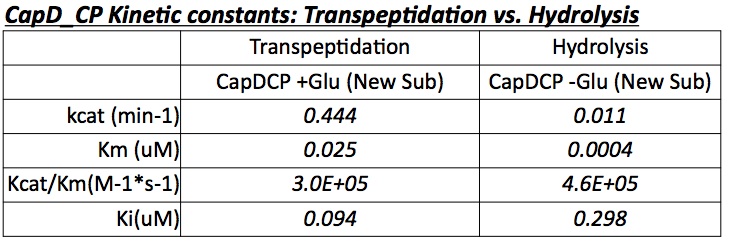
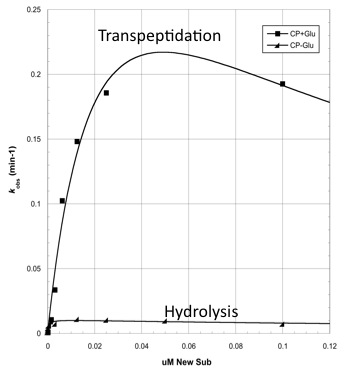
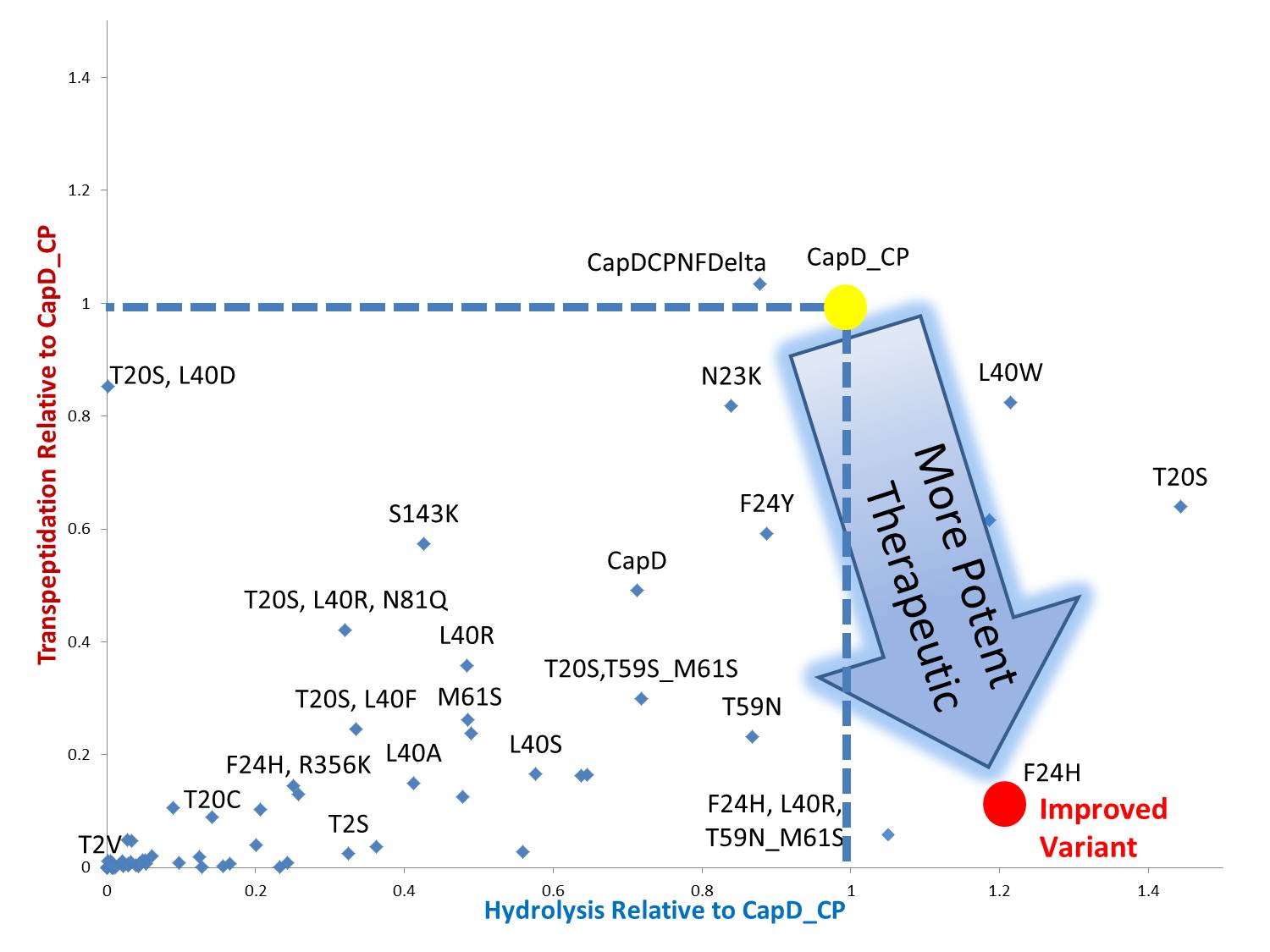
The two abilities of CapD_CP are transpeptidation and hydrolysis. Based on the Kcat and Km (see figure 4) values of the two, we conclude that CapD_CP is a weak binder and efficient catalyst for the transpeptidation reaction. In terms of hydrolysis, it shows strong binding but slow catalysis, thus our mutant designs focus on increasing the catalytic efficiency.
Mutant Designs
By standardizing the activity slope of each design relative to CapD_CP, a scatterplot easily portrays the qualities of each mutant. Several designs show negative catalytic curves similar to the catalytic knockouts. Some immediately show a negative activity curve meaning decrease in transpeptidation, hydrolysis, or both. T20S is a promising mutant hydrolase design.
 "
"