Team:Washington/Gram Positive/Test
From 2010.igem.org
Contents |
Enzyme Assay

Reason
Enzyme assays are commonly used experiments for quantifying enzyme activity by measuring the decrease in substrate or increase in product concentration.
Measuring Medium
By continuously measuring the difference in substrate and product fluorescence emission on a plate reader, our assay can quantify the efficiency of CapD_CP mutants.
Goal
For this project, two conditions are assayed to determine a mutation's change in hydrolytic activity. The two conditions are: L-Glutamate and no L-Glutamate. For the first condition, because L-Glutamate is the secondary substrate which assists in dissociating the covalent linkage of the primary substrate, PDGA, and CapD_CP, by binding to PDGA, thus releasing and helping in regeneration of the CapD_CP enzyme, we are testing the transpeptidation capability of the enzyme. PDGA is translocated and re-attached to a secondary substrate, L-Glutamate. For the second condition, water is used in place of L-Glutamate as the secondary substrate to release the enzyme from PDGA. Water has a low affinity for our enzyme; therefore, the expected result is a slower rate of regeneration, and thus lower overall enzyme activity. The ultimate goal for the project is to make mutant designs which increase the rate of hydrolysis.
Materials
To assimilate the cleavage process occurring in-vitro, we use a lab-safe PDGA that contains a linked fluorophore and quencher. When the link exists, there is no emission due to light illumination from the plate reader because all light energy is absorbed by the quencher. When the PDGA is cleaved by our enzyme, the embedded linked fluorophore and quencher is disrupted, preventing the quencher from absorbing the light energy and allowing light emission which can be measured by the plater reader.
Data Analysis
CapDCP Activity Validation
Before we predict which mutations increase hydrolysis capability, we need to validate that the circularly permuted version of CapD has measurable activity for further assessments. We also hypothesize a threonine residue in the catalytic site of CapD_CP plays an important role in the catalysis reaction and mutating it will eliminate all enzymatic activity. Thus we created two mutants, T2V and T2A, to act as negative controls. The result (figure 1 below) of this assay confirms our hypothesis that CapD_CP has enzymatic activity to the two catalytic knockouts. The relatively flat activity curves of the knockout mutants confirm the hypothesis of the threonine's role in the catalytic site.
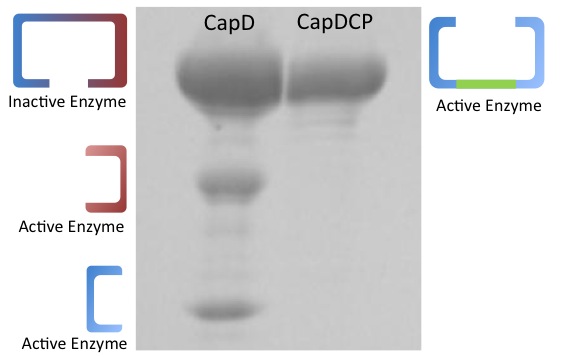
Protein Gel
Before determining the kinectic properties of the two enzyme versions, they were run on an agarose gel (figure 2 on the right) to determine the physical traits. Based on the gel, the two enzymes show different physical properties with CapD producing three bands and CapDCP producing only one. Two of the bands shown for CapD are equal in intensity but different in size. The higher intensity band is the same size as CapDCP's only band.
Continuing with CapD_CP
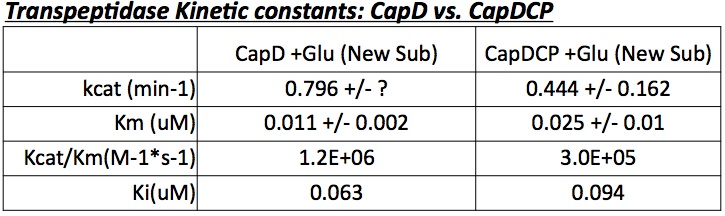
Based on the gel's results, CapD_CP was used for future mutant designs. The reason is two-fold. First, CapDCP is easier to quantify due to its single band characteristic. Second, testing with the three unequal and ambiguous bands in CapD would prove problematic since determining which band to measure for active enzyme is nearly impossible. After validating the activity of CapDCP, more analysis was done pertaining enzymatic properties. Compiling a Michaelis-Menten profile, the data provides properties of Kcat, Km, Kcat/Km and Ki (see figure 3)
for the CapD and CapDCP enzymes. Based on the gel result and the kinetic properties, evidence shows the two versions are very similar, thus all mutants will be done in CapD_CP.
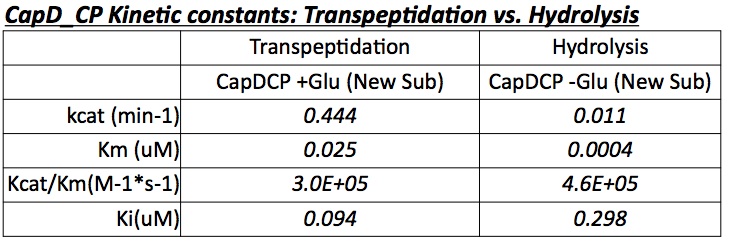
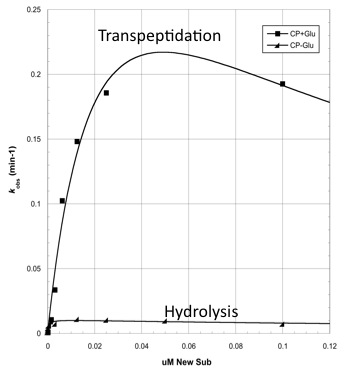
Analyzing CapD_CP
The two abilities of CapD_CP are transpeptidation and hydrolysis. Based on the Kcat and Km (see figure 4)
values of the two, we conclude that CapDCP is a weak binder and efficient catalyst for the transpeptidation reaction. For hydrolysis, it shows strong binding but slow catalysis. Since the goal is an increase in hydrolysis, mutants enhance the hydrolysis reaction of CapDCP. Since CapD_CP is a strong binder in terms of its hydrolysis capability, strong binding CapDCP mutants must be designed.
Mutant Designs
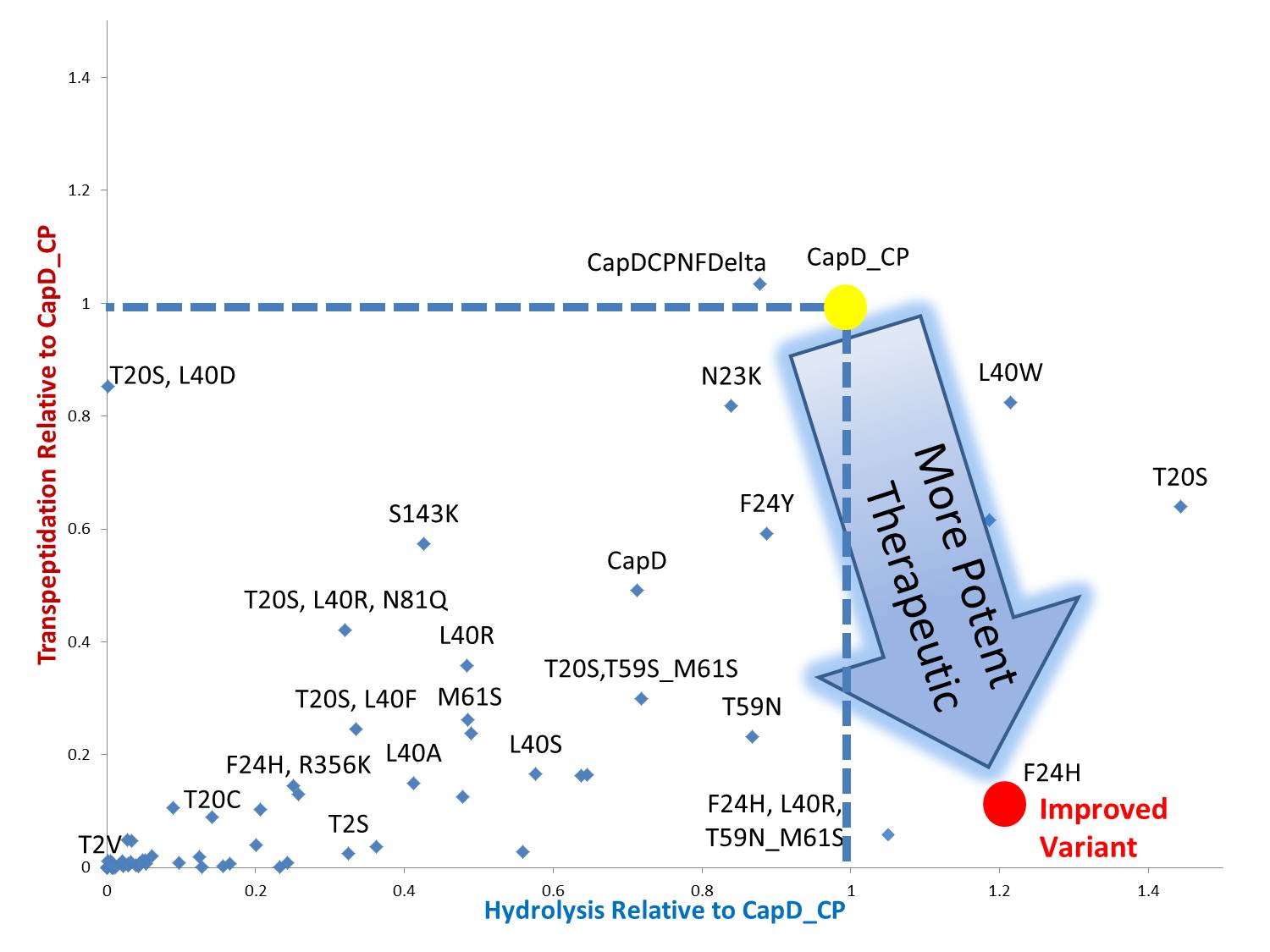
By standardizing the activity slope of each design relative to CapD_CP, a scatterplot easily portrays the qualities of each mutant. Several designs show negative catalytic curves similar to the catalytic knockouts. Some immediately show a negative activity curve meaning decrease in transpeptidation, hydrolysis, or both. T20S is a promising mutant hydrolase design.
 "
"