Team:Washington/Gram Negative/Build
From 2010.igem.org
(→Building the Tse2/Tsi2 Toxin/ Antitoxin system) |
|||
| Line 25: | Line 25: | ||
</html> | </html> | ||
<!---------------------------------------PAGE CONTENT GOES BELOW THIS----------------------------------------> | <!---------------------------------------PAGE CONTENT GOES BELOW THIS----------------------------------------> | ||
| + | =Building a Type VI Dependent Probiotic= | ||
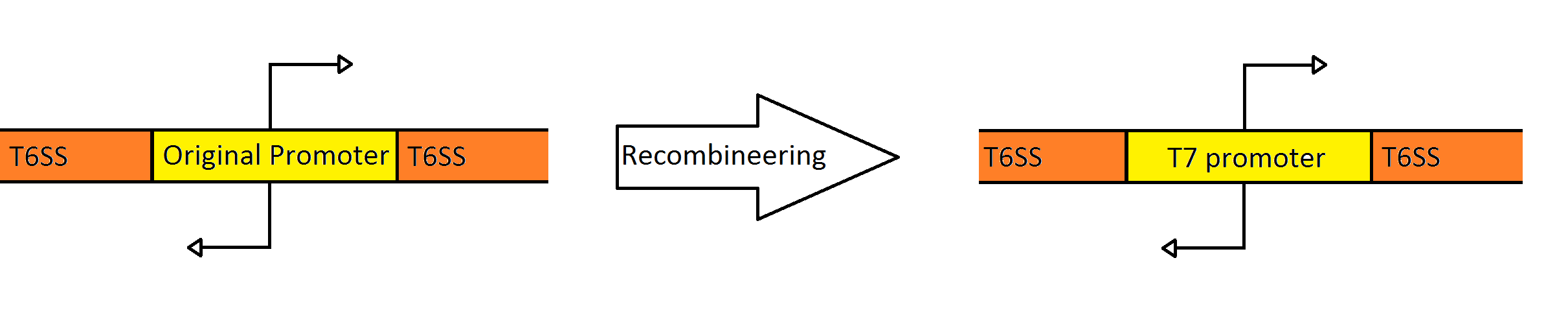
| + | One of the major hurdles of creating our probiotic is the high level of regulation that this system is under in its native host. Several transcriptional and post-transcriptional regulators are present in both promoter regions, which prevented the system from expressing in ''E. coli''. | ||
| - | + | =Creating the Tse2/Tsi2 Toxin/ Antitoxin system= | |
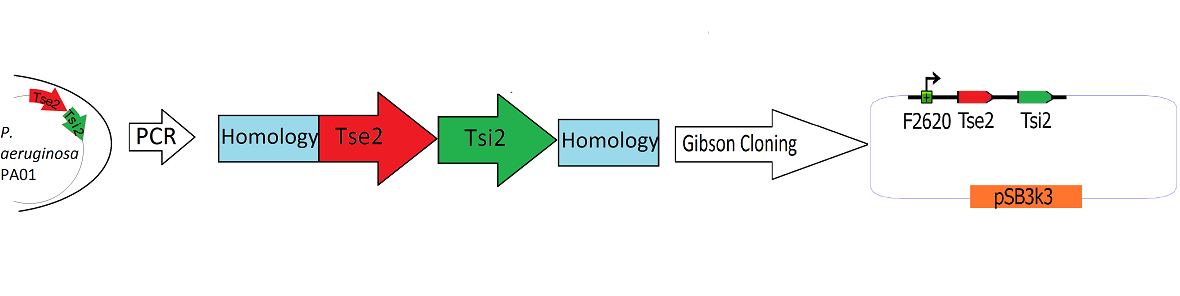
| - | + | The Tse2/Tsi2 locus was amplified using PCR with primers that amplified the region from the start codon of Tse2 to the stop codon of Tsi2.The primers were designed to flank the Tse2/Tsi2 product with approximately 40bp regions of homology to the psb3k3-F2620 plasmid. The forward primer added 44 basepairs of homology to the 3' end of F2620 to the 5' end of the construct. The reverse primer added 38 basepairs of homology to the suffix end of psB3k3 to the 3' end of the construct. The Tse2/Tsi2 locus was then placed downstream from F2620 via Gibson cloning. Gibson cloning is a method that joins regions of homology found on the 5' and 3' ends of linearized molecules (for a more in depth explanation of Gibson cloning, see protocols page[https://2010.igem.org/Team:Washington/Protocols]). The end result of this Gibson cloning reaction was a circularized plasmid containing the final F2620-Tse2/Tsi2 construct in psb3k3. The Gibson reaction mixture was transformed into electrocompetent Dh5a, and the resulting colonies were screened for insert of expected length via double restriction digest followed by agarose gel electrophoresis. The plasmids that showed inserts of expected length were sequenced, and it was determined that multiple plasmids had F2620-Tse2/Tsi2 inserts with expected sequences. | |
| - | = | + | |
| - | The Tse2/Tsi2 locus was amplified using PCR with primers that amplified the region from the start codon of Tse2 to the stop codon of Tsi2.The primers were designed to flank the Tse2/Tsi2 product with | + | |
[[Image:Washington_Building_Tse2_circuit.png|800px|]] | [[Image:Washington_Building_Tse2_circuit.png|800px|]] | ||
Revision as of 03:46, 9 October 2010
Building a Type VI Dependent Probiotic
One of the major hurdles of creating our probiotic is the high level of regulation that this system is under in its native host. Several transcriptional and post-transcriptional regulators are present in both promoter regions, which prevented the system from expressing in E. coli.
Creating the Tse2/Tsi2 Toxin/ Antitoxin system
The Tse2/Tsi2 locus was amplified using PCR with primers that amplified the region from the start codon of Tse2 to the stop codon of Tsi2.The primers were designed to flank the Tse2/Tsi2 product with approximately 40bp regions of homology to the psb3k3-F2620 plasmid. The forward primer added 44 basepairs of homology to the 3' end of F2620 to the 5' end of the construct. The reverse primer added 38 basepairs of homology to the suffix end of psB3k3 to the 3' end of the construct. The Tse2/Tsi2 locus was then placed downstream from F2620 via Gibson cloning. Gibson cloning is a method that joins regions of homology found on the 5' and 3' ends of linearized molecules (for a more in depth explanation of Gibson cloning, see protocols page[1]). The end result of this Gibson cloning reaction was a circularized plasmid containing the final F2620-Tse2/Tsi2 construct in psb3k3. The Gibson reaction mixture was transformed into electrocompetent Dh5a, and the resulting colonies were screened for insert of expected length via double restriction digest followed by agarose gel electrophoresis. The plasmids that showed inserts of expected length were sequenced, and it was determined that multiple plasmids had F2620-Tse2/Tsi2 inserts with expected sequences.
 "
"