Team:USTC Software/Features
From 2010.igem.org
(Difference between revisions)
(New page: {{Team:USTC_Software/Header}} ==Fun and Function== == MoDeL:Modeling Database Language == === Bring complex modeling to the next level === [[Image:Ustcs cnmodel 0.jpg|400px|Figure 1: Chai...) |
(→MoDeL:Modeling Database Language) |
||
| Line 3: | Line 3: | ||
==Fun and Function== | ==Fun and Function== | ||
== MoDeL:Modeling Database Language == | == MoDeL:Modeling Database Language == | ||
| + | <br /> | ||
=== Bring complex modeling to the next level === | === Bring complex modeling to the next level === | ||
| - | + | ---- | |
| - | + | {| cellpadding="10" cellspacing="0" | |
| - | Binding in complex is characterized by connecting Nodes to form trees. It’s as simple as constructing a new node with bond nodes as its children. For example the binding structure of TetR dimer is considered as a TetR2 node with two TetR nodes as its children. Available nodes can be any part on a chain or even nodes of binding sites, which allows you to create a huge binding tree or even a forest of trees. Node makes the description of complicated binding in complex possible. | + | |- |
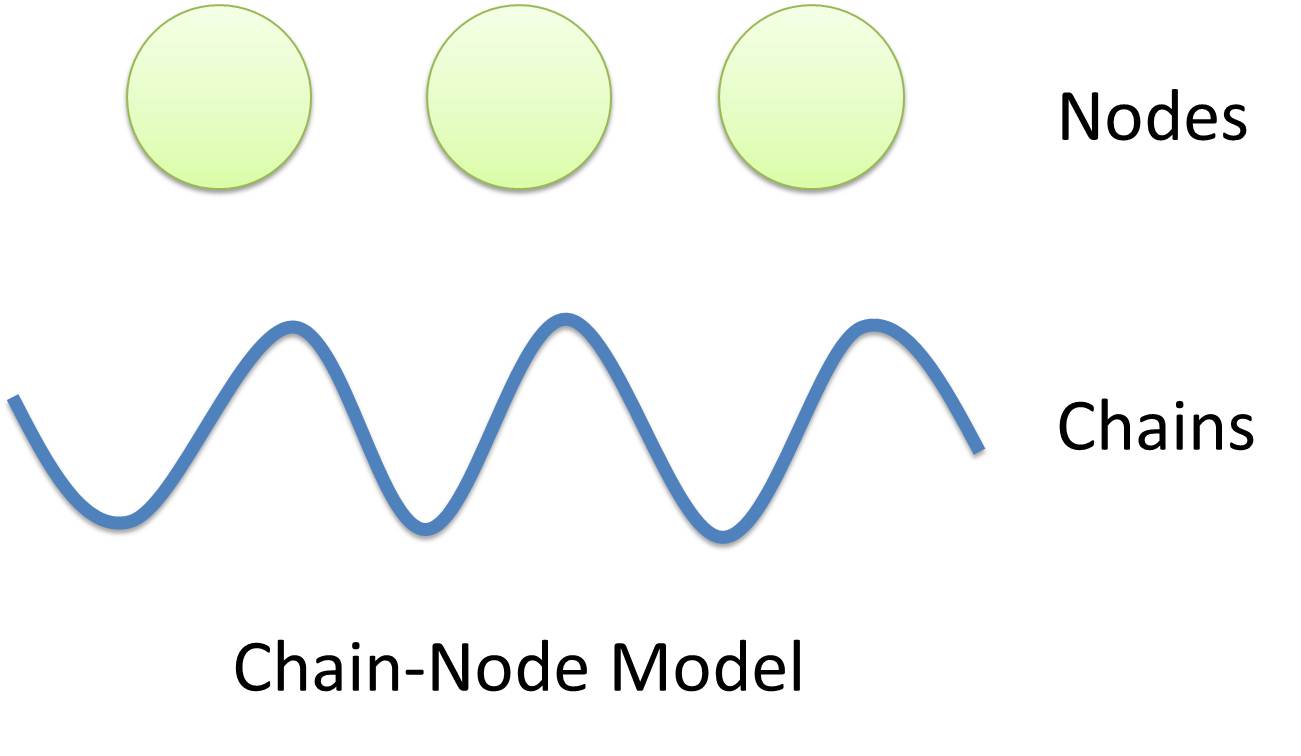
| - | Together with Chains and Nodes, you can almost model any complex you have in mind. So enjoy the freedom of Chain-Node Modeling! You are suggested to read this One- | + | |[https://2010.igem.org/Team:USTC_Software/model_features '''Chain-Node Model'''](Figure 1) is a brand new modeling concept that includes both detailed structure description and universal applicability. Instead of treating complex as a whole, Chain-Node Model view complex as a construction of it basic '''Parts'''. One basic assumption of our model is that complex always carries its parts’ properties. Though imprudent sometimes, this assumption greatly extend the usability of automatic modeling, with just a few parts, one can construct a bunch of complex with functions. |
| - | + | |rowspan="2"|[[Image:Ustcs cnmodel 0.jpg|thumb|400px|Figure 1: Logo of Chain-Node Model]] | |
| + | |- | ||
| + | |Binding in complex is characterized by connecting '''Nodes''' to form trees. It’s as simple as constructing a new node with bond nodes as its children. For example the binding structure of TetR dimer is considered as a TetR2 node with two TetR nodes as its children. Available nodes can be any part on a chain or even nodes of binding sites, which allows you to create a huge binding tree or even a forest of trees. Node makes the description of complicated binding in complex possible. | ||
| + | |- | ||
| + | |colspan="2"|Together with Chains and Nodes, you can almost model any complex you have in mind. So enjoy the freedom of Chain-Node Modeling! You are suggested to read this [https://2010.igem.org/Team:USTC_Software/MoDeL One-Minute Introduction] to have an intuitive idea of our modeling system. | ||
| + | |} | ||
| + | <br /> | ||
=== Modeling with templates === | === Modeling with templates === | ||
| - | + | ---- | |
| - | Similar reactions happen for similar reasons. For example, the repression of pTetR promoter by TetR dimer can happen on many different DNA molecules with pTetR, regardless of what other biobricks is constructed on the DNA. With this thought, we add the part of Substituent, which can replace inactive parts of Species in Reactions, making it more general, and actually turning it into a reaction template. Having substituent, a pTetR TetR binding reaction is re-interpreted as the binding reaction of ANY | + | {| cellpadding="10" cellspacing="0" |
| - | + | |- | |
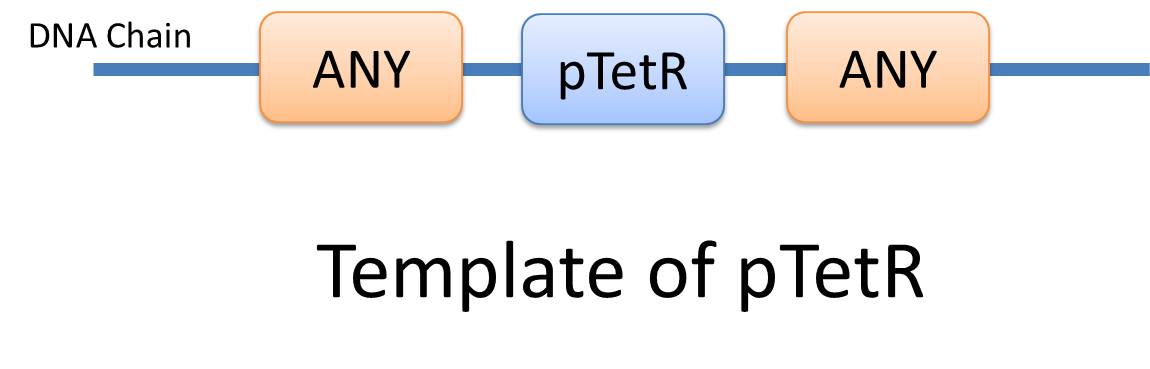
| + | |Similar reactions happen for similar reasons. For example, the repression of pTetR promoter by TetR dimer can happen on many different DNA molecules with pTetR, regardless of what other biobricks is constructed on the DNA. With this thought, we add the part of [https://2010.igem.org/Team:USTC_Software/model_features '''Substituent'''], which can replace inactive parts of Species in Reactions, making it more general, and actually turning it into a reaction template. Having substituent, a pTetR TetR binding reaction is re-interpreted as the binding reaction of '''ANY''' DNA with pTetR(Figure 2) with ANY protein with TetR-dimer binding site. Modeling with templates allows us to describe reactions of new complex even without rewrite the reactions and species in database. | ||
| + | |[[Image:Ustcs Template ptetr.jpg|thumb|400px|Figure 2: Template of pTetR DNA]] | ||
| + | |} | ||
| + | <br /> | ||
=== Automatic modeling database language === | === Automatic modeling database language === | ||
| - | + | ---- | |
| - | We use a database to store all the information we need in modeling. In order to realize automatic modeling, we construct the database in unified format and make it machine-readable. Every component of database has its specified attributes and values, which makes the format of the database a unique yet standard database language. We call it MoDeL: | + | {| cellpadding="10" cellspacing="0" |
| + | |- | ||
| + | |We use a database to store all the information we need in modeling. In order to realize automatic modeling, we construct the database in unified format and make it machine-readable. Every component of database has its specified attributes and values, which makes the format of the database a unique yet standard database language. We call it [https://2010.igem.org/Team:USTC_Software/model_features '''MoDeL''']''': Mo'''deling '''D'''atabas'''e''' '''L'''anguage. MoDeL is based on XML language, which makes it flexible and extensible. For more specifications of MoDeL, click [https://2010.igem.org/Team:USTC_Software/model_lang here]. | ||
| + | |rowspan="5"|[[Image:Ustcs dblang.jpg|thumb|400px|Figure 3: A peek at our database]] | ||
| + | |- | ||
| + | |<br style="clear:both;"> | ||
| + | |- | ||
| + | |<br style="clear:both;"> | ||
| + | |- | ||
| + | |<br style="clear:both;"> | ||
| + | |- | ||
| + | |<br style="clear:both;"> | ||
| + | |} | ||
==Auto-Modeling Algorithm== | ==Auto-Modeling Algorithm== | ||
Revision as of 16:59, 16 October 2010
Contents |
Fun and Function
MoDeL:Modeling Database Language
Bring complex modeling to the next level
| Chain-Node Model(Figure 1) is a brand new modeling concept that includes both detailed structure description and universal applicability. Instead of treating complex as a whole, Chain-Node Model view complex as a construction of it basic Parts. One basic assumption of our model is that complex always carries its parts’ properties. Though imprudent sometimes, this assumption greatly extend the usability of automatic modeling, with just a few parts, one can construct a bunch of complex with functions. | |
| Binding in complex is characterized by connecting Nodes to form trees. It’s as simple as constructing a new node with bond nodes as its children. For example the binding structure of TetR dimer is considered as a TetR2 node with two TetR nodes as its children. Available nodes can be any part on a chain or even nodes of binding sites, which allows you to create a huge binding tree or even a forest of trees. Node makes the description of complicated binding in complex possible. | |
| Together with Chains and Nodes, you can almost model any complex you have in mind. So enjoy the freedom of Chain-Node Modeling! You are suggested to read this One-Minute Introduction to have an intuitive idea of our modeling system. | |
Modeling with templates
| Similar reactions happen for similar reasons. For example, the repression of pTetR promoter by TetR dimer can happen on many different DNA molecules with pTetR, regardless of what other biobricks is constructed on the DNA. With this thought, we add the part of Substituent, which can replace inactive parts of Species in Reactions, making it more general, and actually turning it into a reaction template. Having substituent, a pTetR TetR binding reaction is re-interpreted as the binding reaction of ANY DNA with pTetR(Figure 2) with ANY protein with TetR-dimer binding site. Modeling with templates allows us to describe reactions of new complex even without rewrite the reactions and species in database. |
Automatic modeling database language
| We use a database to store all the information we need in modeling. In order to realize automatic modeling, we construct the database in unified format and make it machine-readable. Every component of database has its specified attributes and values, which makes the format of the database a unique yet standard database language. We call it MoDeL: Modeling Database Language. MoDeL is based on XML language, which makes it flexible and extensible. For more specifications of MoDeL, click here. | |
 "
"