Team:UCL London/Project Modeling
From 2010.igem.org
PROJECT HYPOXON - MODELLING
“The process defines the product” is the motto we always have in mind when designing the route for the production of a compound.
Biopharmaceuticals are commonly synthesized using E. coli as production chassis. Typically, the production of biopharmaceuticals is triggered by the addition of an induction agent, often IPTG, during the bioprocess. Imagine if the bacteria as we know it today could drive protein expression by linking the hypoxia condition to oxygen sensitive promoters within the cells. Synthetic biology through biochemical engineering can create the potential at which Escherichia coli cells- and we all know how harmful to humans can specific strains be- used in a twisted yet beautifully way as an expression system for the production of biopharmaceuticals. The project is based on the engineering modification of the cells and the construction of a self-induced biological circuit that allows an automated protein expression.
We all remember the Swine flu pandemic that outbreak that occurred about a year ago. The production of such a great amount of pharmaceuticals in such a short time to cover the needs of the infected people was only based on a well planned engineering scale-up equations and an established protocol. Therefore, in order to create a product and with regards to our project, a pharmaceutical drug, there is the need to establish the conditions that will enable the effective development of the process and the efficient and most economically viable product. By decreasing the cost of the project, it only means that it is delivered to the patient at a lower cost. By producing naturally occurring drugs at a large scale, avoiding as much as possible chemicals that can cause damage to the cells or side-effects of the final product it only means that a safer product has been manufactured and it is ready for consumption by the patient.
Biological engineering is yet indeed in the early years of its life- if it would be an innovative product it would be on the introduction phase, where there is around 2% of innovators and followers of that particular idea. What about the rest though? How do we make it possible for others to understand and moreover how do we make the whole idea approachable to others? This project had to be translated into the actual benefits that the society could gain from this. Therefore, as more “mature” engineering fields exist like mechanical and electrical, the project was “expressed” in those terms. For example the standard computational programmes SolidWorks and Verilog are powerful tools for the expression of equipment and circuits respectively in the engineering fields mentioned above. Comparatively engineering toolboxes that will enable the visual construction of models, using those programmes for the needs of a synthetic biology, will enable a quantitative understanding of how parts of a biological system when come together behave in different manners. Through this combination it is believed that the whole perspective of Synthetic Biology will be transformed (Carlson, 2009). The construction of a database with well characterised genetic parts, with assigned mathematically abstracted functions that can be modelled in a software and turning electronic circuits into molecule representations via DNA synthesis could lead to a more economically and social value of what Synthetic Biology projects can offer. The currently available computational tools for the representation of genetic networks, include Antimony, Athena, BioJade, GenoCAD, OptCircuit, SynBioSS and many more software tools which were constructed to integrate all the relevant aspects of a biological system (Purnick and Weiss, 2009).
According to Carlson (2010), the revenues from genetic modifications of biological systems in USA back in 2007 was around 2% of the GDP (gross domestic product). These products included drugs, crops, materials, industrial enzymes and fuels. The Parliament Office of Science and Technology (2008) posted that the global market for DNA sequencing technology and services, in 2006, as estimated by the US Department of Energy was over $7 billion. This rapid revenue growth from the sales of biotechnological products, suggests the evolution of a new market. The necessity to improve current biological technologies and aid the advancement of establishing them is still an issue. Large scale projects have demonstrated social and economic implications, such as the GM crops and the political, social and ethical implications it raised as a result of the introduction of Genetic Modifications as a new technological approach (Gregory and Jay Lock, 2008). The challenges, when it comes to the complexity of the biological systems and the application of them in our life are still influenced by the public policy, safety and biosecurity which suggest that although Biology is technology, and is in fact the oldest technology, Synthetic Biology is only at the beginning. Based on the uncertainty that some features in biology carry, the optimisation of such algorithms is still not reliable. For example, E.coli is a quite simple biological organism and is used as a chassis for a wide range of biological expressions; however, it still contains many uncharacterized genes (Carlson, 2010).
One of the goals of this project is to be able to take it from bench top to commercialisation of the technology and to optimise the conditions at a pilot scale prior to moving to large quantities production, at low cost and at higher yield. This could mean access to medication for people who could not have otherwise afforded it. In fact, many affected families live with less than $1 a day leading to the main question: How can these people get treatment since they can only spend a few pennies a day on drugs and the per capita health budget is only a few US dollars per year? (Carlson, 2010) According to WHO (World Health Organisation) the disease harms whole society by decreasing the GDP of the country. In particular (Gallup and Sachs, 2001) malaria affected countries reduces GDP growth by 1.3% per year while WHO reported that malaria-free countries have a threefold higher GDP per capita compared to those countries. From synthetic biology to biochemical engineering and from the genetic modifications of an E.Coli cell to the growth of that cell in order to achieve protein expression we aim to build a synthetic project where several parts come together and create a protocol for the production of pharmaceuticals. That’s not the whole story though. Our aim goes further into the modification of bacteria and accordingly other systems such as yeast or mammalian cell culture, the establishment of the optimum conditions to achieve their growth and expression of the desired product, downstream processing that should be adjusted to fit the needs of each system with the final goal to scale-up the process to cover the needs of large populations. What if, through an organised and well pictured project, where all the parts, like LEGOS, fitted together in all the possible ways through the synthetic biology game and one day we could come across to that specific gene modification that will enable the fight of cancer or hepatitis or even HIV? What if? Just imagine that.
We tried to predict the models that would enable such visualization with the hope that one day there will be specific genetic engineering software with its own database of biobricks and tools used for the random or algorithmic construction of biological parts that when assembled in reality can create a better biological system than already existed.
“It is only through learning how the parts work, and how they work together, that we will truly be able to produce engineered biological systems” Robert H. Carlson
The aim of adopting modelling tools is to enable us to make right predictions with regards to our project and to develop a standard protocol to be used as the basis for translating the construction of a biological circuit such as the bacterial (E.coli) circuit, to other expression systems for the production of novel biopharmaceuticals for the treatment of major diseases.
The modelling section is divided into 5 main sections for simplicity: 1. Circuit Construction
2. Bioprocess Flowsheet Development
3. Fermentation set-up: Mechanical Design of the Fermenter & Materials
4. Large-scale considerations: Economic Evaluation, Health & Safety Analysis, Waste Management & HAZOP studies
CIRCUIT DESIGN
Many thanks to Dr. Yuhong Zhou for all her help in developing the mathematical equations
The design of a biological system should include all the information needed to describe what is happening within the cell (Carlson, 2010). As in with our case, the numerical simulation of all the biochemical reactions identified, was to describe the system, and specific values such as the rate constant, are unknown, leading to the simulation exploring a wide variety of possible values from literature. Even if we did have particular values for the variables appearing in the model, it would still be impossible to make accurate predictions about its behaviour in real life. According to Carslon (2010), the line between model and simulation becomes blurred when the only way to assess a particular model is depending upon the simulation of the model. However, again the design stage with the simulation differs significantly from the actual model. This is where the difficulty lays, as scientists and engineers, work on projects together by repeatedly comparing simulation and measurement through experiments. The Bristol team for this year iGEM competition, has assisted us, in the production of a standard simulation, with the potential to run multiple simulations using their BioSim programme, to explore the behaviour of the model, over a range of conditions. However, the experiments, in terms of the laboratory experiments of the physical system, to explore the model behaviour, must be carried out and hopefully with the aid of theoretical behavioural descriptions from textbooks, we can produce quantitative predictions.
The SyntheticBiology.org website defines Synthetic Biology as:
A) the design and construction of new biological parts, devices, and systems, and
B) the re-design of existing, natural biological systems for useful purposes.
The cartoon description used to describe the protein expression as a result of the transcription of DNA into RNA by polymerase which is in turn translated into protein. The ribosomes, which are not included in the animation, play an important role in the production of the proteins, according to the “central dogma”, defined by Francis Crick in 1957 that information flows from DNA to RNA to protein.
The challenges around the biological systems are the following, and summarise the scope of the formation of the iGEM competition:
1. Defining the device physics of molecular components 2. Building and simulating models 3. Using defined components from a standard library (biobricks) 4. Building new biological entities based on quantitatively predictive designs
The genetic switch, that we aimed to build in the bacterium E.coli, was to be independent of the application of a chemical signal, such as IPTG, that would induce the cells to start producing the protein. The bacterium, when the switch is on, is predicted to make red fluorescent protein; when the switch is off, the protein is no longer made and any remaining protein decays.
The fundamental that the genetic circuit works on, is that a signal from outside the cell, that is when the oxygen spike appears, declaring the low oxygen concentration present, induces the cell to begin producing the proteins. Traditionally, this was done with the manual addition by the operator of IPTG to the cells. The employment of inducible promoters, i.e. pNark, that works by interfering with a repressor protein, LacI, a protein that inhibits transcription by binding to the promoter. The polymerase, T7RNA P, is located downstream of the pNark, where it binds to DNA and begins transcription. The repressor protein, LacI, blocks the access to the pLacI promoter, because it is prevented by the pNark promoter. Therefore the role of the pNark inducible promoter is double: to accept the signal for cell induction and production of mRNA and proteins through the loop and also to inhibit the repressor binding to DNA. The addition of IPTG into the system, will cause the induction of the pLacI promoter which was repressed by the LacI proteins during the duration of the positive feedback loop and will be the regulator. The anti-PA binds to the promoter activator and reversibly stops the positive loop and hence, transcription terminates.
According to Purnick and Weiss (2009), the first wave of synthetic biology includes the combination of promoters, RBS (ribosome binding sites) and transcriptional repressors for the formation of functional modules e.g. controlled protein expression. Such a novel circuit design was based on principles like the creation and analysis of a genetic circuit, the experimental evaluation of the circuit and the combatibility of the results with literature. Moreover, they refer to directed evolution, which is about the optimisation of biobricks and the system i.e. the biological circuit.
BIOPROCESS FLOWSHEET
Many thanks to Zarah Ali for all her help in the production of the SuperPro flowsheets.
According to Makrides (1996) the periplasm of E.coli contains only about 4% of the total cell protein. The periplasmic expression allows expression of fragments to be effectively concentratd. Other reports (Humphreys, 2004) that the antibody expression of Fab fragments can take place in the periplasm of E.coli and purified with an aqueous periplasmic heat extraction, which eliminates most of the host cytoplasmic and membrane proteins, followed by ion exchange chromatography.
E. coli is an established production system of choice for antibody fragments used in therapeutic applications. One reason is that E. coli provides the means to progress from antibody selection to Good Manufacturing Practice (GMP) production of antibodies in a rapid manner. The other reason is the fact that high production levels of antibody fragments are attainable when using E. coli. As of 2004, there have been several Fabs or Fab’s in clinical trials run by Genentech and Celltech, where the fragments have been produced using E. coli (Anderson et al., 2004).
Fermentative Pathways
Host: Escherichia Coli
Advantages:
Provides a wide choice of cloning vectors Easily controlled gene expression Gives large yields Secretes good protein Provides fast growth rate
Disadvantages:
Lacks post-translational modifications Posses high levels of endotoxins Forms inclusion bodies (i.e. protein aggregates)
Escherichia coli produces antibody fragments rather than whole antibodies due to the fact that it lacks post-translational modifications and also since polymeric polypeptide assembly is not well supported (Johansson, 2007).
FERMENTER MECHANICAL DESIGN
Special thanks to Achilleas Constantinou from the Department of Chemical Engineering for the COMSOL Multiphysics fermentation simulation
Mechanical Design of 1000L Fermenter
This report was conducted to estimate the dimensions and the materials needed for the construction of a 1000L fermenter for the production of Fab fragments (fragmented antigen binding) are the regions of an antibody expressed and secreted in E.coli cells. For the estimation of the dimensions of the fermenter empirical equations and rules of thumbs were exploited. The dimensions and the fermenter properties like the diameter, the height as well as the impellers speed, the baffles position and the driveshaft size were estimated on a scale- down model. The material used for the fabrication of the fermenter, the head end, the ports and the pipes is stainless steel to resist corrosion and maintain a clean and sterile vessel. The diameter of the vessel is 0.8m and the height is 2m. The thickness of the wall is 6mm with a 0.15 m jacket surround it. The impeller’s speed is 3.3 m/s and the impeller used is Ruston Turbine four blades, suitable for the growth of E.coli cells. Therefore all the power requirements were satisfied ensuring the viability of the design. Five spray balls were found to be enough for a sufficient cleaning of the fermenter with their 360° coverage and placed on the inside of the torispherical head in a cross shape. Then the flowrate in the pipes was predicted enabling the calculation of the diameter. Basically all the information specified for every single part of the fermenter was taken from recommendations, advice from experts, theoretical predictions as well as engineering justifications. Last but not least emphasis is given on the importance of maintenance and annual shutdowns of the plant as well as regular checks to ensure its proper operation.
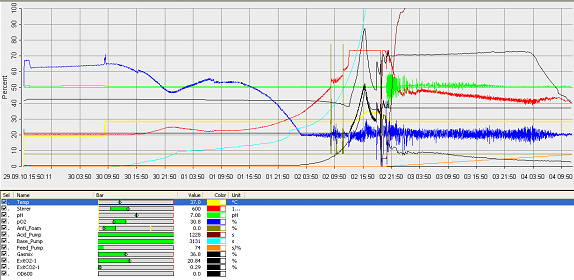
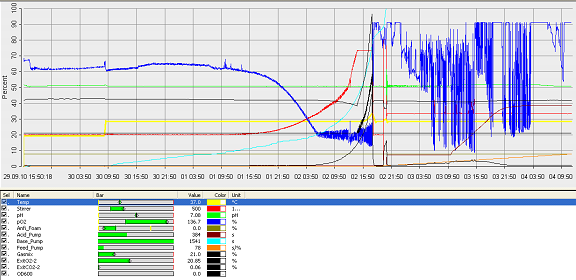
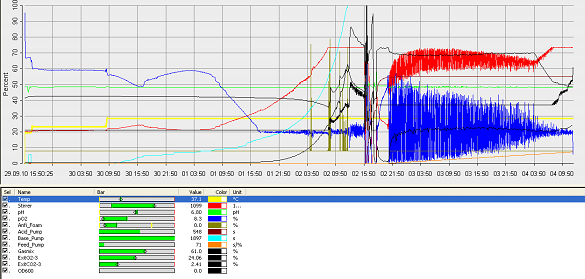
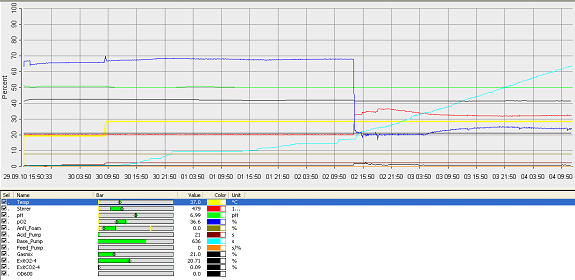
- fermenter graph***
and since the vessel is metal, the value of the thickness means it would aid cleaning. Operation Cycle
The operation cycle for the fermenter involves start up, where process water is fed from the process water port and media is fed from the feed port into the vessel. The level of the liquid is measured using the level indicator probe. Once the required level is met then the ports close. Air is pumped through the air inlet to the sparger inside the fermenter.
The temperature is brought and maintained to the optimum temperature. There are two probes in the fermenter which are at the greatest range i.e. opposite sides of the fermenter as shown in the drawing. This is to ensure the distribution of heat is even and the average of both sensors should read 37°C. The pH is maintained at a desired value by the addition of acid and alkali. Two probes for measuring the pH are placed inside the fermenter at opposite ends to ensure there is good mixing as this would be indicated if there was little change between the two probes.
Once the optimum conditions have been achieved the innocculum is fed through the innocculum port. Antifoam from the antifoam port is added to avoid foam formation which reduces the efficiency of the process. The exhaust gases are released from the fermenter whilst in operation and the pressure is maintained. The cells are grown for a certain period of time and the process is regulated using a series of automatic control loops.
Finally there is shut down, where the fermentation batch ends and in this step all the broth is collected into the harvest port.
After each operation cycle the fermenter will have to be cleaned and sterilized in place. CIP will be performed using acid and base detergents which will be pumped through the cleaning solution ports into the 3 spray balls. SIP requires that the fermenter and all pipes are sterilized with steam at 121°C. The steam will come from the acid, base and antifoam ports. Regular and annual maintenance checks will have to be performed to ensure everything is running correctly and as efficiently as possible.
18.3 Maintenance
Typically, 3-5% of fermentations in an industrial plant are lost due to failure of sterilisation procedures. However in antibiotic fermentations like in this project, fewer than 2% of production scale antibiotic fermentations are lost through contamination by microorganisms or phage. (Doran, 1999)
Industrial fermenters are designed for in situ steam sterilisation under pressure. For effective sterilisation, all air in the vessel and pipe connections must be displaced by steam. After sterilisation, all nutrient medium and air entering the fermenter must be sterile.
Maintenance is required to ensure the fermentation process is achieving its goals, to identify any problems that may occur and ultimately keep the process working as efficiently as possible to save on costs. Maintaining the fermenter can significantly reduce the overall operating cost as well as minimise the risk of any hazards. For maintenance, the top of the fermenter can be removed to allow access of maintenance personnel for any repairs that may be required. For large vessels a domed construction at the top to allow access to personnel is less expensive and so this has been chosen in the design (Doran, 1999).
Once the design has been built and approved by British Standards 5500, the fermenter can then start to operate (Sinott, 1999). Maintenance is crucial and the fermenter must be monitored at all times. Most measurements can be made on-line through probes in the vessel which are sensors that detect changes in many variables such as temperature and pH. However, whilst monitoring some values there may always be a delay. For example, in a typical fermentation, the time scale for change in pH and dissolved-oxygen tension is several minutes to make sure they are all correctly calibrated and functioning properly and as efficiently as possible. This is to avoid any problems that may be caused because of a false reading. For example if the pH probe is not working properly and the pH is either too high or too low, then the cells would die and also the product may denature. Also possible blockages in pipes or broken valves must be checked regularly. Oxygen supply through bottom sparging should also be checked regularly, as cells die without it and that would mean a significant loss in time and money if the fermenter has been running for several days.
The fermenter requires annual shutdown to check for possible corrosion on the internal surface and also on the impellers. Corrosion may reduce the speed and efficiency of them impellers and the shaft too. This needs to be maintained at a high standard to ensure good mixing in the fermenter for growth of cells and product. Baffles also need to be checked for to see the level of corrosion. Also there may be possible fouling in the jackets and general piping problems, such as blockages and leakages, as well as overall contamination in the system. As previously mentioned in the design a 2mm allowance for corrosion had been added on the thickness of all equipment. The annual check would see how much all the equipment has been eroded and identify any problems to resolve them.
The aim of the Mechanical Drawing is to provide a detailed analysis of the construction and operation of the fermenter that is a technical way of presenting this piece of equipment. In this section the dimensions and materials of construction of a 1L fermenter were estimated. The fermenter is used for the growth of Escherichia coli cell culture for the production of therapeutic fragmented antibodies.
For the design project purpose several assumptions had to be made. The first one was the model taken for the scale- up process. This was the original 1L fermenter used for the pilot scale of the E.coli cells. Assuming that a 1000L fermenter operating with 75% space efficiency (0.75) and 1 marine impeller (di=dv/2). The vessel aspect ratio is 2.5 that is Ah/d= 2.5 and the fermenter is aerated at 0.75 vvm (that is 0.75 volumes of air per volume of liquid per minute). The gassed power requirement per m3 is Pg/VL= 1860 W/m3 as it was found from the model used for the scale- up. Usually the industrial motors are used to deliver 1-3 W/L (Lye et Baganz, 2007). Therefore the 1.86 W/L was chosen on the assumption that due to engineering cell modifications, the E.coli cells are not so robust and thus extremely high power inputs should be avoided. For this purpose instead of using a marine propeller a lighter choice was preferred since the Chinese Ovary cells are susceptible to mechanical damage (Farid, 2006) due to their large size and lack of rigid cell wall. Therefore the power input value must be kept to a minimum. A Rushton turbine four blade impeller is used as the axial flow type of impeller ensuring medium speed and ensuring medium growth rate but also good mixing.
19.2 Calculations for determination of the fermenter’s properties
The operating pressure of the vessel was assumed to be 3.5 bar and the design pressure 3.85 bar. The operating temperature for mammalian cells is 37°C (Lydersen et al, 1994) but the design temperature is 4- 121°C allowing the introduction of steam to clean the vessel. Since the mammalian cells are very sensitive, an elevation in temperature e.g. 40°C can cause cell damage and death. Hence temperature probes allowing 0.5°C from the temperature set point as well as pH probe to maintain the appropriate range was considered.
Power Requirement Calculation
The gassed power input Pg is 30- 40% from the ungassed value, Pug. That is due to a reduction in density due to the mixture of the gas and the liquid phase compared to the ungassed liquid only. Also that is due to the formation of cavities which reduce the drag on the impeller, reducing the power required to rotate it at a given speed (Farid, 2006). Starting by assuming Pg= 0.4 Pug Pg= * VL With Pg usually between 1-3 W/L, so Pg/ VL= 1860 W/m3 (small scale) and need to find the properties of the 1000L fermenter: VV= 1000L= 1m3 VL= 0.75* 1m3= 0.75m3
The fermenter is aerated at 0.75vvm, i.e. 0.75 volumes of air per volume of liquid per min Then Pg= 1860 W/m3 * 0.75m3 which is the total gassed power input for the 1000L fermenter Pg= 1395W Then Pug= = 3488W The aspect ratio of the vessel height to the vessel diameter could be from 1-3 and according to experts (Jacobs) for mammalian cells the value of 2 is advicable. Ah/d= 2.5 Then used:
So hv= 2.5* 0.8m= 2m The minimum thickness required can be calculated from the equation With f= 135N/mm2 for 150°C (121°C actually)
Pi= 3.5 bar= 3.5* 105 Pa, Di= 0.8m and f= 135*106 N/m2 Then e= 1.03*10-3 m ≈ 0.001 m or 1mm
The minimum practical wall thickness of the vessel though should be 2mm. The vessel diameter’s is 0.8m and for vessels of 1m diameter the minimum thickness should be 5mm (Sinnott, 2000).
Jacket
The space between the jacket and the vessel is estimated based on the size of the jacket. According to Sinnott (2000) the size of the jacket is 50mm for small vessels and 300 mm for large vessels. The assumption made is that the jacket’s size should be: 150 mm for the 1000L fermenter.
The jacket surrounding the fermenter will be made out of stainless steel 304 and its thickness is the same as the fermenter. The wall thickness of the fermenter was estimated to be 6mm based on rules of thumb including an additional 2mm allowance for corrosion.
Impeller size calculation
Then the diameter of axial mixers was based on the empirical statement (Lye, 2006) that is over 0.33 to 0.5 the tank diameter. So assumed to be 0.45 timed the diameter of the vessel, that is dm= 0.45* 0.8m= 0.36 m and the spacing is usually 1-2 impeller diameters of 1 tank diameter (Lye, 2006). So spacing, based on 1.5 * dT=0.36 *0.8m= 0.3m impeller spacing of one impeller from the other.
Impeller Speed and Reynolds number estimation
The flow of air is 0.75 vvm that is Qair= (m3/s) So Qair= (0.75m3 * 0.75)/ 60 s Qair= 0.00935 m3/s≈ 0.01m3/s Then in order to find N use the Power number Po: Po= 0.9 * (5.2 *1)
So Po= 4.68 N= Assuming the density and viscosity properties of the bacterial cell culture are similar to those of water (non- viscous fluid): Escherichia coli: rod shaped cells, 3μm and 1μm diameter, ρ= 1000 kg/m3 and μ= 0.01 Pa *s N= 1.31 s-1 or 79rpm Then utip= πdiN= 3.3 m/s Re= = 210713Turbulent flow
Driveshaft & Blades Size calculation
Then the impeller driveshaft was decided to be bottom driven as this is recommended in small industrial scale production where the shaft is shorter, gives better power development leaving more space on top plate for ancillaries with only one drawback of a more difficult seal arrangement (Lye et Baganz, 2007).
The shaft diameter was calculated by the inequality: ds The shaft length was assumed to be 1.65 m (slightly above the liquid’s level). f is the design stress which is taken from literature which is 175 N/mm2 for stainless steel 316L working at 37°C. The shaft diameter found is 0.07 m. Then the blade thickness: dbt where n is the number of impellers, so n=1 dbt=0.002 m Blade width= 0.2* di= 0.16m Blade height= 0.25* di= 0.20m Disc diameter= 0.75* di= 0.60m Disc thickness= Blade thickness= 0.02m
Baffles
It was decided that four diametrically opposed baffles would be put in the vessel. The size of each baffle should be 0.1- 0.05 tank diameter. The assumption made here is that the baffle’s diameter= 0.1*dT= 0.1 *0.8m= 0.08m or 8cm. The baffle height is dependent on the gas holding or it accepted to be equivalent to the cylindrical height of the vessel. In this case it was assumed to be slightly above the liquid’s level. Small gap between vessel and baffle is recommended to stop biomass build up, enhance oxygen transfer and cleaning solution (Lye et Baganz, 2007). The gap is 0.1 of the baffle width fixed to the vessel such that they can be removed. Also it is recommended that baffles are placed at an angle as with were found from experts to enhance scouring (Lye et Baganz, 2007). The dimensions of the baffles were estimated based on empirical equations relating their size to the vessel’s size.
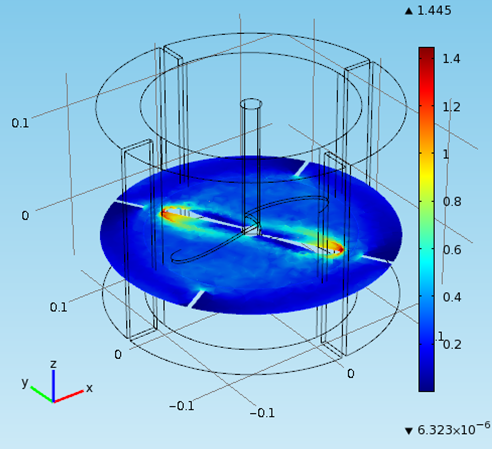
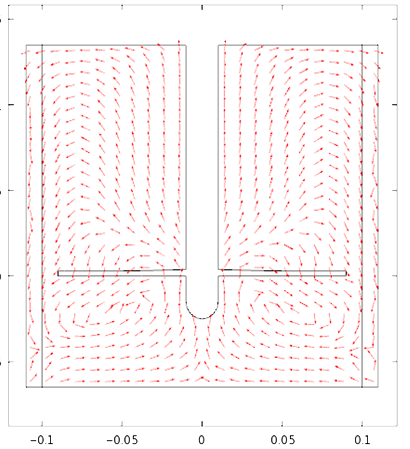
Therefore in the mechanical drawing the baffles were drawn at an angle of 10° to the vertical pointing anti- clockwise since the flow direction is clockwise. That was decided after some thought and with the preparation of a computational fluid dynamic model to predict the flow of the fluid. Also the base of the cylindrical vessel was chosen to be rounded at the edges since in this way it discourages the formation of stagnant regions.
Orifice Sparger
An orifice sparger was placed under the bottom impeller and its size was calculated to be 75% the impellers diameter that is 3.375 cm (4 cm for easier design). The holed on this circular ring must not exceed 6 mm and therefore for design calculations their size was assumed to be 5 mm. A magnetic drive was chosen as the driveshaft seal as this is more common in non- viscous fermentation broths like mammalian cell culture which is a very dilute culture since the cell density is low. Although since it is placed at the bottom of the fermenter can cause cellular disruption it is an easy seal as separate parts and the shaft does not penetrate the vessel. It is made up from one driving magnet and one driven magnet and it is clearly seen on the drawing.
Design of closure for the fermenter
It was decided to use a standard torispherical head since it is widely used in industry and can withstand operating pressure up to 15 bar. The bending and shear stresses were taken into consideration in the design of the heads. For the estimation of these stresses usually caused by dilations of the vessel a basic equation used for a hemisphere was used. The knuckle and crown radii were found from empirical values allowing the stress concentration value determination that allowed with its turn the estimation of the thickness of the head.
Cs=
where Cs is the stress concentration factor taken to allow for the increased stress due to the discontinuity. Rc= crown radius (not to be more than the vessel’s diameter) Rk= knuckle radius The ratio Rc/ Rk should not be less than 6% (to avoid buckling).
e= For formed heads (no joints in the head) J=1.0
Pi is the internal pressure and f is the design stress (literature value). The design pressure= 110% of the operating pressure (Coulson and Richardson, 2000) In this case, the fermenter is operating at 3.5 bar therefore the design temperature is 3.85 bar. Operating temperature= 37°C mammalian cell culture, cleaning solution 4°C for the cooling water and 121°C for the steam (CIP) f= 135 N/mm2 (stainless steel corresponding to operating temperature 150°C) Rc= 1.5 m hence Rk= 0.06*Rc So Rk=0.09 m So e= 7 mm (5mm+ 2mm for corrosion allowance) For design purposes it was assumed that the wall thickness and the end thickness are the same, 6mm. Basically, the wall thickness has an impact on the overall heat transfer coefficient while the end thickness affects the mixing characteristics of liquid inside and since the vessel is metal, the value of the thickness means it would aid cleaning.
LARGE SCALE CONSIDERATIONS
ECONOMICAL CONSIDERATIONS
HEALTH & SAFETY ANALYSIS
WASTE MANAGEMENT
HAZARD STUDIES
Fermentation


 "
"



 Twitter
Twitter Facebook
Facebook UCL
UCL Flickr
Flickr YouTube
YouTube