Team:UCL London/Introduction
From 2010.igem.org
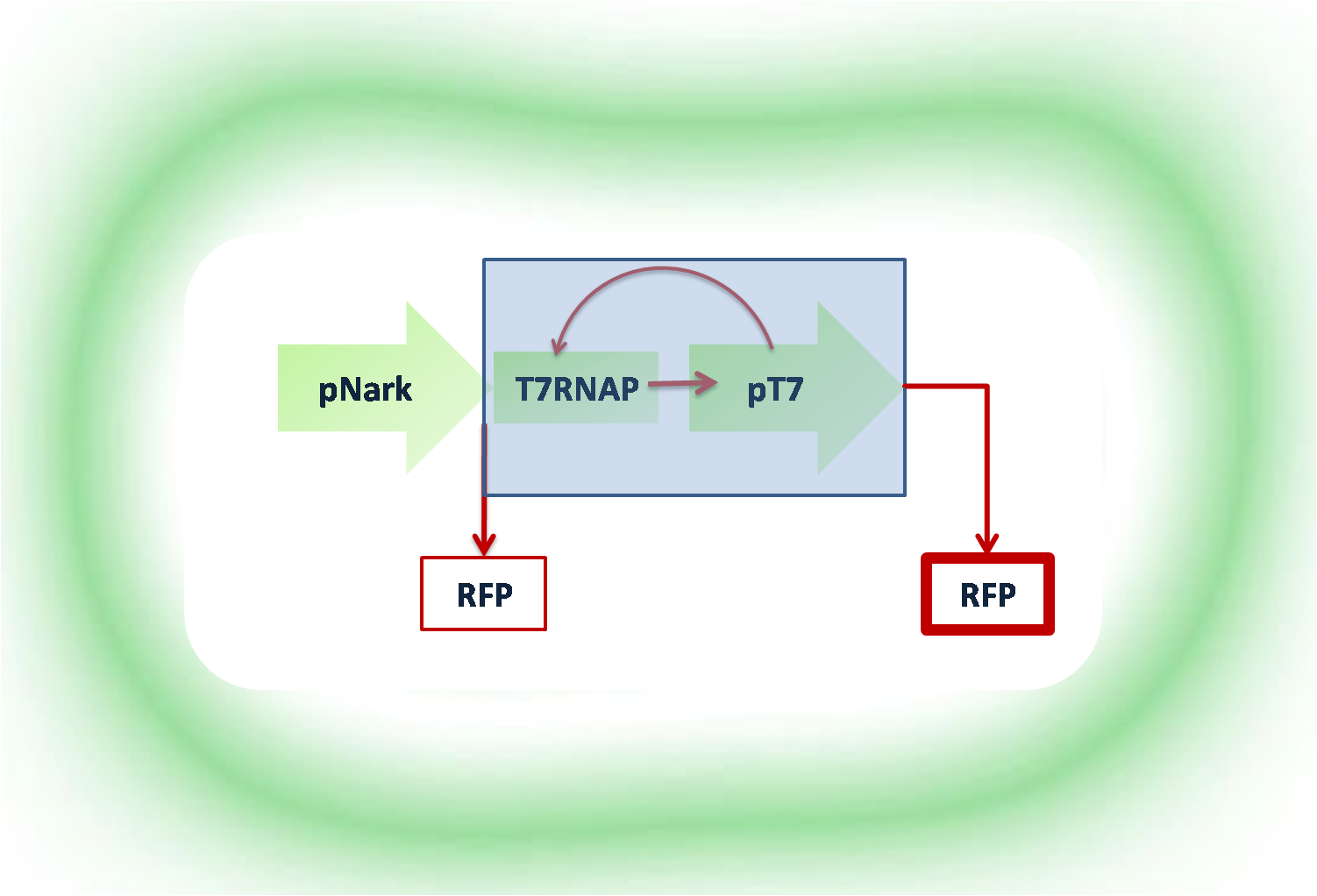
Project Hypoxon Modelling Introduction
The process defines the product” is the motto we always have in mind when designing the route for the production of a compound.
Biopharmaceuticals are commonly synthesized using E. coli a production chassis. Typically, the production of biopharmaceuticals is triggered by the addition of an induction agent, often IPTG, during the bioprocess. Think of it as the chocolate cake where milk (the oxygen in our case for the cells) is added. The cells consume all the milk (oxygen) to grow and then they are ready to start producing the desired protein. Until now, in industry this was achieved by manually inserting a chemical inducible agent, IPTG, which stimulates the cells to start making protein. In the case of the chocolate cake, you need the milk (for a different reason) to make a nice homogeneous broth and cook it until the liquid evaporates and the cake is indeed baked.
The system we are proposing, which theoretically through modelling, has been proven to work, shows that when there is no milk-oxygen- a signal linked to the cells, through the biological circuit enables their auto-induction. Therefore, in the traditional cake baking, instead of manually stirring the milk with the rest of the ingredients and once it is ready, you make a thick broth and you cook it. Imagine now, like the thermomixer, where you put everything together and stir, and when the time is right, the cake is cooked. Similarly, at the time when the oxygen is consumed, and a spike is shown on the fermentation panel, notifying the operator (or the chef) about it, there will be no need for the any extra chemical, as through the "Pavlovian Principle" it was proven that the cells will realise this through a series of signas from the biological circuit and start producing the protein in a more natural manner.
The modelling section is divided into 5 main sections for simplicity and you can see each by clicking on them:
We all remember the Swine flu pandemic outbreak that occurred about a year ago. The production of such a great amount of pharmaceuticals in such a short time to cover the needs of the infected people was only based on well planned engineering scale-up equations and an established protocol. Therefore, in order to create a product and with regards to our project, a pharmaceutical drug, there is the need to establish the conditions that will enable the effective development of the process and the efficient and most economically viable product. By decreasing the cost of the project, it only means that it is delivered to the patient at a lower cost. By producing naturally occurring drugs at a large scale, avoiding chemicals that can cause damage to the cells or side-effects of the final product only means that a safer product has been manufactured and it is ready for consumption by the patient.
“It is only through learning how the parts work, and how they work together, that we will truly be able to produce engineered biological systems” Robert H. Carlson
The aim of adopting modelling tools is to enable us to make right predictions with regards to our project and to develop a standard protocol to be used as the basis for translating the construction of a biological circuit such as the bacterial (E.coli) circuit, to other expression systems for the production of novel biopharmaceuticals for the treatment of major diseases. Our collaboration simulates the performance of our circuit, based on our equations, to predict the models that would enable such visualization with the hope that one day there will be specific genetic engineering software with its own database of biobricks and tools used for the random or algorithmic construction of biological parts that when assembled in reality can create a better biological system than already existed. Could BSim do that? how the BSim modelling program works Bristol BSim
From synthetic biology to biochemical engineering and from the genetic modifications of an E.coli cell to the growth of that cell in order to achieve protein expression we aim to build a synthetic project where several parts come together and create a protocol for the production of pharmaceuticals. That’s not the whole story though. Our aim goes further into the modification of bacteria and accordingly other systems such as yeast or mammalian cell culture, the establishment of the optimum conditions to achieve their growth and expression of the desired product, downstream processing that should be adjusted to fit the needs of each system with the final goal to scale-up the process to cover the needs of large populations. What if, through an organised and well pictured project, where all the parts, like LEGOS, fitted together in all the possible ways through the synthetic biology game, one day we could come across to that specific gene modification that will enable the fight of cancer or hepatitis or even HIV? What if? Just imagine that.
Biological engineering is in the early years of its life- if it would be an innovative product it would be in the introduction phase, where there is around 2% of innovators and followers of that particular idea. What about the rest though? How do we make it possible for others to understand and moreover how do we make the whole idea approachable to others? This project had to be translated into the actual benefits that the society could gain from this. According to Carlson (2010), the revenues from genetic modifications of biological systems in USA back in 2007 was around 2% of the GDP (gross domestic product). These products included drugs, crops, materials, industrial enzymes and fuels. The Parliament Office of Science and Technology (2008) posted that the global market for DNA sequencing technology and services, in 2006, as estimated by the US Department of Energy was over $7 billion. This rapid revenue growth from the sales of biotechnological products, suggests the evolution of a new market. The necessity to improve current biological technologies and aid the advancement of establishing them is still an issue. Large scale projects have demonstrated social and economic implications, such as the GM crops and the political, social and ethical implications it raised as a result of the introduction of Genetic Modifications as a new technological approach (Gregory and Jay Lock, 2008).
The challenges, when it comes to the complexity of the biological systems and the application of them in our life are still influenced by the public policy, safety and biosecurity which suggest that although Biology is technology, and is in fact the oldest technology, Synthetic Biology is only at the beginning. Based on the uncertainty that some features in biology carry the optimisation of such algorithms is still not reliable. For example, E.coli is a quite simple biological organism and is used as a chassis for a wide range of biological expressions; however, it still contains many uncharacterized genes (Carlson, 2010). One of the goals of this project is to be able to take it from the bench shelf to commercialisation of the technology and to optimise the conditions at a pilot scale prior to moving to large quantities production, at low cost and at higher yield. This could mean access to medication for people who could not have otherwise afforded it. In fact, many affected families live with less than $1 a day leading to the main question: How can these people get treatment since they can only spend a few pennies a day on drugs and the per capita health budget is only a few US dollars per year? (Carlson, 2010) According to WHO (World Health Organisation) the disease harms the whole society by decreasing the GDP of the country. In particular (Gallup and Sachs, 2001) malaria affected countries reduces GDP growth by 1.3% per year while WHO reported that malaria-free countries have a threefold higher GDP per capita compared to those countries.


 "
"




 Twitter
Twitter Facebook
Facebook UCL
UCL Flickr
Flickr YouTube
YouTube