Team:UCL London/Characterisation
From 2010.igem.org
(→Fermenter 1 - UCL_London 2009 iGEM BBa_K239011 PART) |
|||
| (61 intermediate revisions not shown) | |||
| Line 54: | Line 54: | ||
='''1. CHARACTERISATION OF NEW PARTS'''= | ='''1. CHARACTERISATION OF NEW PARTS'''= | ||
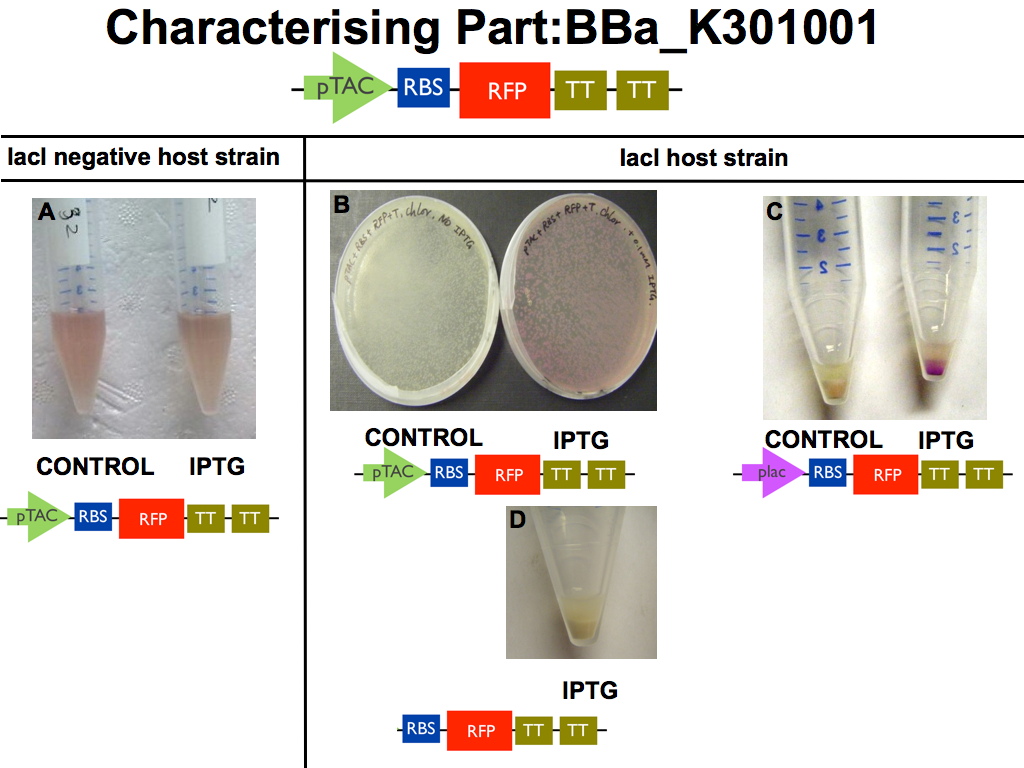
| - | [ | + | Our pTAC with RFP[http://partsregistry.org/wiki/index.php?title=Part:BBa_K301001 BBa_K301001] reporter was characterised at lab scale by carrying out a successful transformation with a control. |
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[Image:Ucl-Part-BBa-K301001.png|center|800px]] | ||
{{:Team:UCL_London/clear}} | {{:Team:UCL_London/clear}} | ||
| - | |||
| - | |||
| - | |||
| + | '''IPTG induction of the reporter device BBa_K301001 was demonstrated in a lacI host strain with 0.1mM IPTG (panel B). A lac promoter device was tested as positive control (panel C).''' | ||
| - | + | '''As predicted, IPTG induction was not observed when our new device resided in a lacI negative host strain (panel A) or when the pTac biobrick was omitted (panel D).''' | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | The following is the manual for the construction of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K301000 BBa_K301000] | |
| - | + | ||
| - | + | ||
| + | <html> <div style="width:100%;overflow: hidden"><img src="https://static.igem.org/mediawiki/2010/7/79/UCL-Constructing-Part-BBa-K.png" width="1000px" style="margin:auto" /></div></html> | ||
| + | Check out our parts page to see the parts submitted in more detail [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2010&group=UCL_London&Done=1|'''UCL_London 2010 iGEM Team Parts''']. | ||
='''2. CHARACTERISATION OF EXISTING PARTS/DEVICES AT 1 L FERMENTER SCALE'''= | ='''2. CHARACTERISATION OF EXISTING PARTS/DEVICES AT 1 L FERMENTER SCALE'''= | ||
| - | As part of the Gold criteria which is our target, we had to characterise an existing part, and so we decided to characterise 3 devices from our 2009 parts; | + | '''As part of the Gold criteria which is our target, we had to characterise an existing part, and so we decided to characterise 3 devices from our 2009 parts;''' |
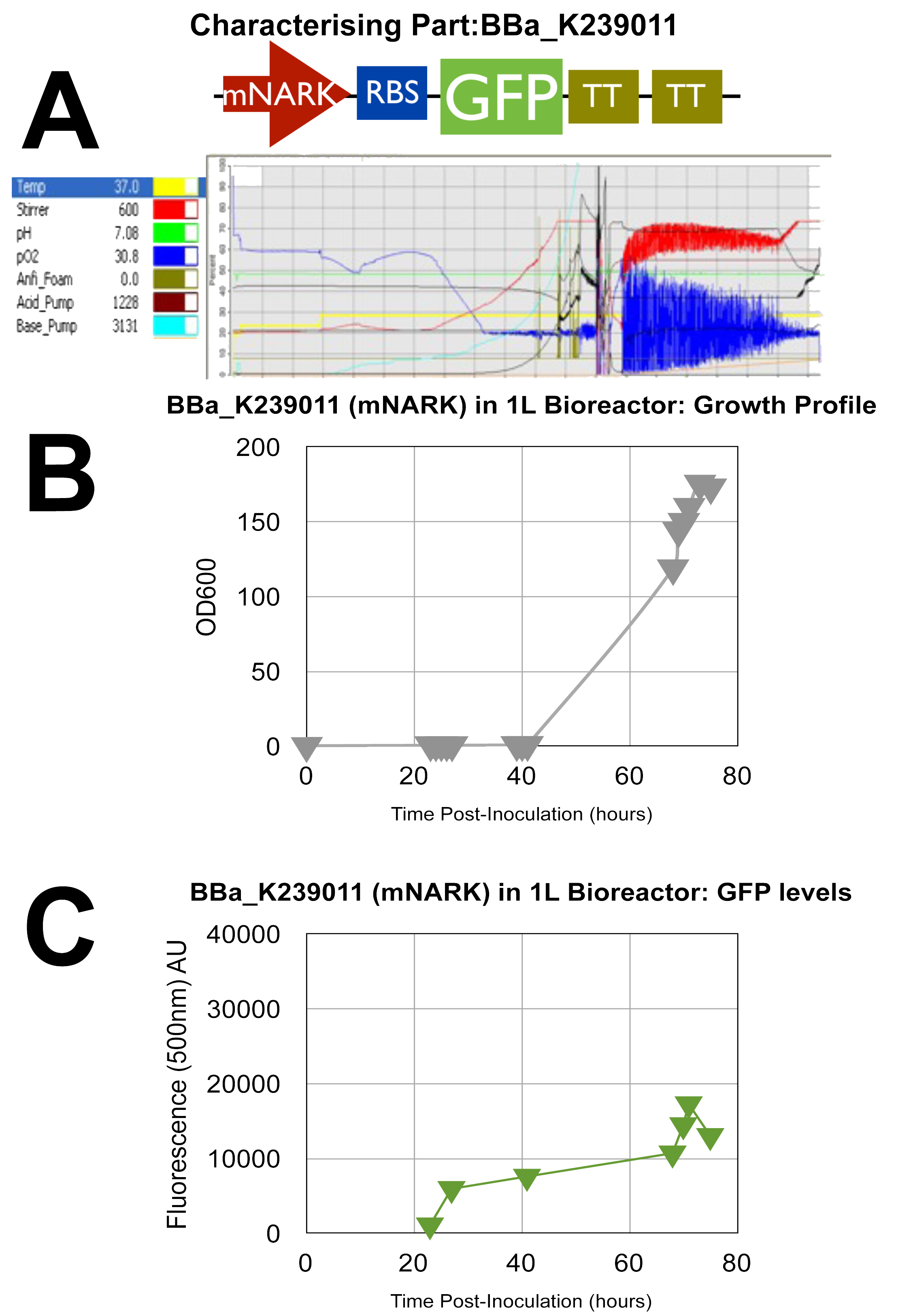
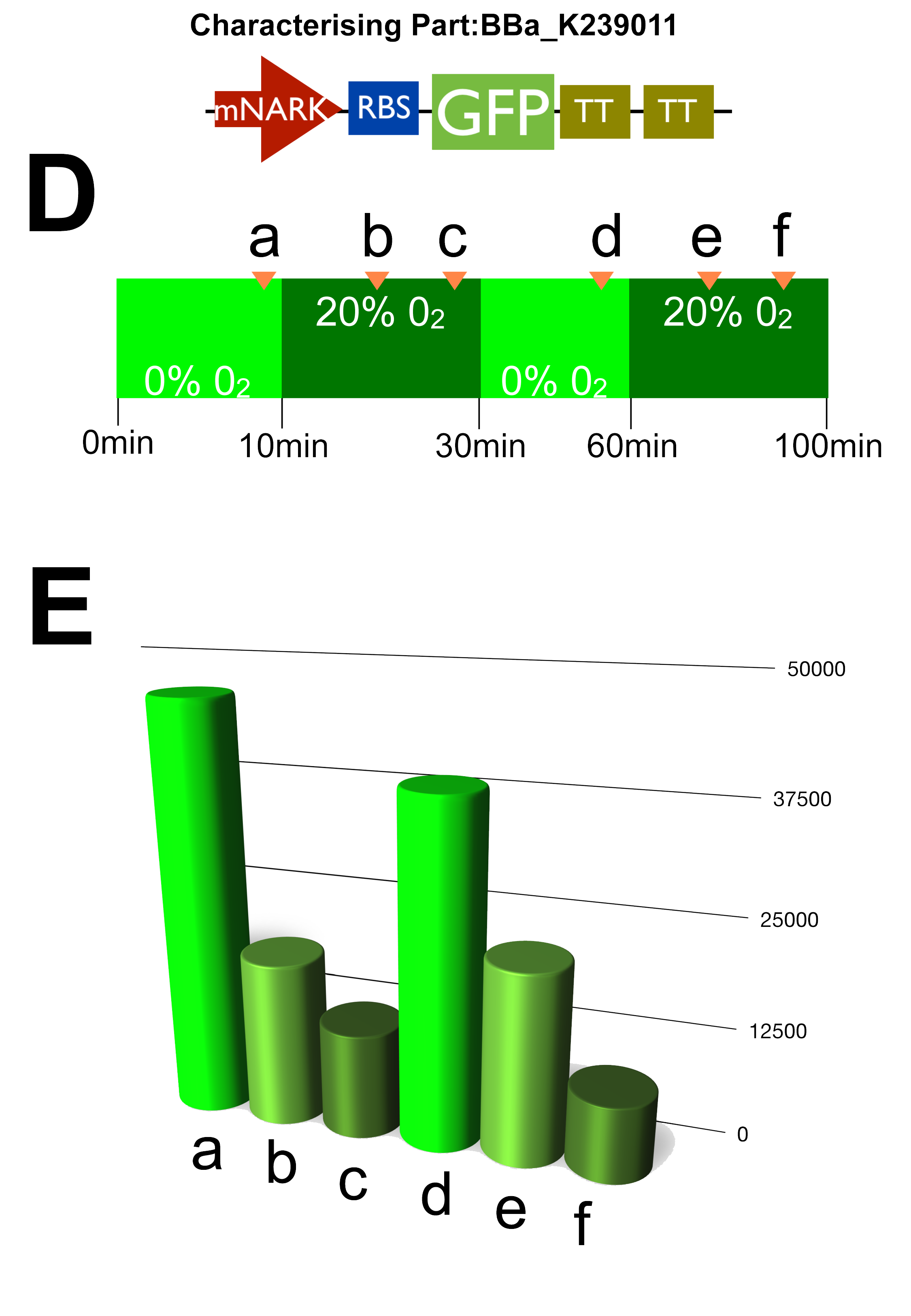
| - | ===Fermenter 1 - [http://partsregistry.org/wiki/index.php?title=Part: | + | ===Fermenter 1 - [http://partsregistry.org/wiki/index.php?title=Part:BBa_K239011 '''UCL_London 2009 iGEM BBa_K239011 PART''']=== |
| - | GFP | + | Stress Light GFP Anaerobic Metabolism Detector: mNARK promoter |
| - | [[Image:UCL- | + | [[Image:UCL-CHARAC11-1.png|800px|left]] |
| + | [[Image:UCL-CHARAC11-2.png|800px|left]] | ||
| - | |||
| - | |||
| - | + | {{:Team:UCL_London/clear}} | |
| + | By analysing the above data, it can clearly be shown that the cells are growing well as they are making use of the oxygen as indicated by the blue oxygen line on A. B indicates their succesful growth showing OD values gradually increasing up to a peak of around 175, although the time taken to do so is approxiamtely double that of normal processes, and this can be attributed to the introduction of new parts into the '''E.coli''' genome resulting in some sort of interaction, that's our theory! | ||
| + | Graph C once again confirms our characterisation as it shows the AU of the green fluorescent dye produced increasing with time. D & E show how the circuit reacts to changes in oxygen at certain intervals where we cut the oxygen of to observe the circuits reaction. | ||
===Fermenter 2 -[http://partsregistry.org/wiki/index.php?title=Part:BBa_K239009 '''UCL_London 2009 iGEM BBa_K239009 PART''']=== | ===Fermenter 2 -[http://partsregistry.org/wiki/index.php?title=Part:BBa_K239009 '''UCL_London 2009 iGEM BBa_K239009 PART''']=== | ||
| - | GFP test devise for | + | GFP test devise for Spy promoter |
| - | + | [[Image:UCL-CHARAC9.png|1000px]] | |
| - | [[Image:UCL- | + | |
... | ... | ||
| - | |||
| + | {{:Team:UCL_London/clear}} | ||
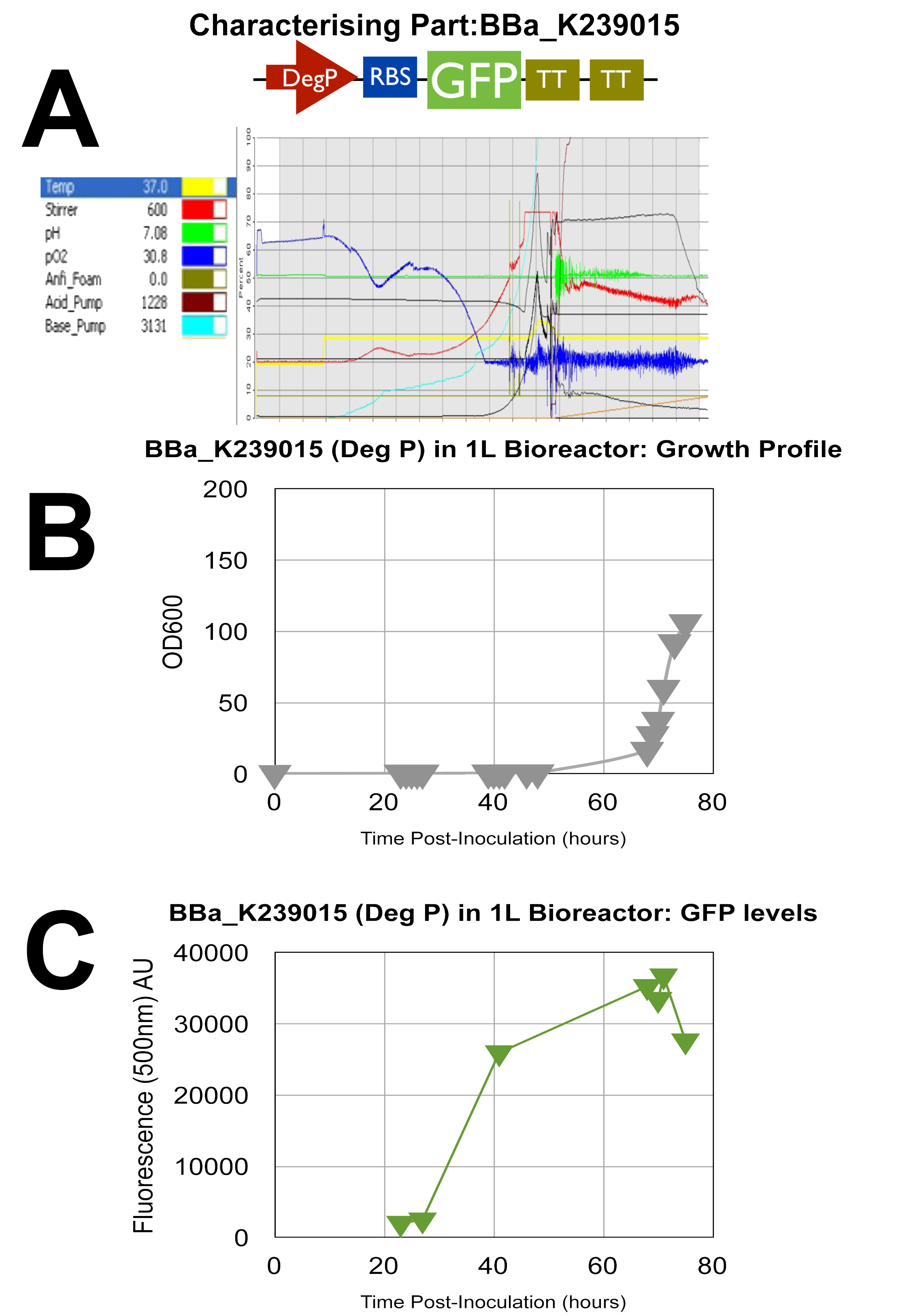
| + | ===Fermenter 3 - [http://partsregistry.org/wiki/index.php?title=Part:BBa_K239015'''UCL_London 2009 iGEM BBa_K239015 PART''']=== | ||
| + | GFP test devise for Deg P promoter | ||
| + | [[Image:UCL-CHARAC15.png|800px|left]] | ||
| Line 125: | Line 120: | ||
| + | {{:Team:UCL_London/clear}} | ||
| - | + | Once again, all 3 graphs show successful characterisation results as the green dye AU is at relatively high levels and the oxygen in A as stated previously is low towards the end of the cycle indicating efficient use of oxygen by the cells. | |
| - | + | ||
| - | + | ||
| - | + | '''''Images from fermentation chamber''''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ''''' | + | |
[[Image:MNARK Infors 05.jpg|300px|left]] [[Image:MNARK Infors 02.jpg|300px|left]] | [[Image:MNARK Infors 05.jpg|300px|left]] [[Image:MNARK Infors 02.jpg|300px|left]] | ||
{{:Team:UCL_London/clear}} | {{:Team:UCL_London/clear}} | ||
| + | |||
| + | The image on the left is that of the control, while that on the right showing the characeterised mNark promoter fluorescent green. | ||
The following video shows the moment of that our iGEM aspirations came true, and we saw green fluorescence; | The following video shows the moment of that our iGEM aspirations came true, and we saw green fluorescence; | ||
| - | + | <html><div style="margin auto"> | |
| - | + | <object width="480" height="420"><param name="movie" value="http://www.youtube.com/v/M0OcVWFOshs?fs=1&hl=en_US&color1=0x234900&color2=0x4e9e00"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/M0OcVWFOshs?fs=1&hl=en_US&color1=0x234900&color2=0x4e9e00" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="480" height="420"></embed></object> | |
| - | <html> | + | </div> |
| - | <object width="480" height=" | + | |
| - | + | ||
</html> | </html> | ||
| + | From the paste removed from the fermenter vessel following the successful characterisation of the parts, we used it to creatively mark the words '''UCL iGEM''' in plates and hence further emphasizing the results that the parts do work. | ||
| + | [[Image:UCL-MNARK_paste.jpg|400px|center]] | ||
| + | {{:Team:UCL_London/clear}} | ||
| + | [[Image:MNARK IGEM plus control 02.jpg|400px|right]][[Image:MNARK IGEM plus control 03.jpg|400px|left]] | ||
| + | {{:Team:UCL_London/clear}} | ||
| + | These images further conclude that we succesfully characterised our mNark Promoter as both images have both a control with no observed dye and one showing the green dye, with which we crafted the words UCL and iGEM as shown above. | ||
| + | ==3. Charactisation of Cambridge Part:BBa_K274003== | ||
| + | [[Image:K274003 Diagnostic Gel .jpg|right|400px]] | ||
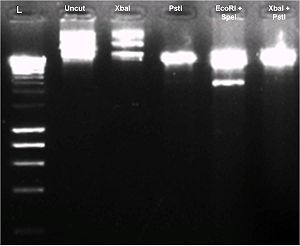
| + | E. coli turned dark green after transformation of this plasmid. This is unexpected as there is no promoter upstream of the operon. | ||
| + | Diagnostic DNA gel was run to test all 4 restriction sites. BBa_K274003 was cut with XbaI, PstI, EcoRI & SpeI, XbaI & PstI. A gel picture is shown as below. | ||
| + | This plasmid did not cut with Xba I restriction site in our hands. As such we could not use it to assemble after other parts. | ||
| - | + | Click on the following link for the [http://partsregistry.org/Part:BBa_K274003:Experience#User_Reviews '''Cambridge Part Review'''] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{{:Team:UCL_London/clear}} | {{:Team:UCL_London/clear}} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Latest revision as of 03:55, 28 October 2010
1. CHARACTERISATION OF NEW PARTS
Our pTAC with RFP[http://partsregistry.org/wiki/index.php?title=Part:BBa_K301001 BBa_K301001] reporter was characterised at lab scale by carrying out a successful transformation with a control.
IPTG induction of the reporter device BBa_K301001 was demonstrated in a lacI host strain with 0.1mM IPTG (panel B). A lac promoter device was tested as positive control (panel C).
As predicted, IPTG induction was not observed when our new device resided in a lacI negative host strain (panel A) or when the pTac biobrick was omitted (panel D).
The following is the manual for the construction of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K301000 BBa_K301000]
Check out our parts page to see the parts submitted in more detail [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2010&group=UCL_London&Done=1|UCL_London 2010 iGEM Team Parts].
2. CHARACTERISATION OF EXISTING PARTS/DEVICES AT 1 L FERMENTER SCALE
As part of the Gold criteria which is our target, we had to characterise an existing part, and so we decided to characterise 3 devices from our 2009 parts;
Fermenter 1 - [http://partsregistry.org/wiki/index.php?title=Part:BBa_K239011 UCL_London 2009 iGEM BBa_K239011 PART]
Stress Light GFP Anaerobic Metabolism Detector: mNARK promoter
By analysing the above data, it can clearly be shown that the cells are growing well as they are making use of the oxygen as indicated by the blue oxygen line on A. B indicates their succesful growth showing OD values gradually increasing up to a peak of around 175, although the time taken to do so is approxiamtely double that of normal processes, and this can be attributed to the introduction of new parts into the E.coli genome resulting in some sort of interaction, that's our theory!
Graph C once again confirms our characterisation as it shows the AU of the green fluorescent dye produced increasing with time. D & E show how the circuit reacts to changes in oxygen at certain intervals where we cut the oxygen of to observe the circuits reaction.
Fermenter 2 -[http://partsregistry.org/wiki/index.php?title=Part:BBa_K239009 UCL_London 2009 iGEM BBa_K239009 PART]
GFP test devise for Spy promoter
Fermenter 3 - [http://partsregistry.org/wiki/index.php?title=Part:BBa_K239015UCL_London 2009 iGEM BBa_K239015 PART]
GFP test devise for Deg P promoter
Once again, all 3 graphs show successful characterisation results as the green dye AU is at relatively high levels and the oxygen in A as stated previously is low towards the end of the cycle indicating efficient use of oxygen by the cells.
Images from fermentation chamber
The image on the left is that of the control, while that on the right showing the characeterised mNark promoter fluorescent green.
The following video shows the moment of that our iGEM aspirations came true, and we saw green fluorescence;
From the paste removed from the fermenter vessel following the successful characterisation of the parts, we used it to creatively mark the words UCL iGEM in plates and hence further emphasizing the results that the parts do work.
These images further conclude that we succesfully characterised our mNark Promoter as both images have both a control with no observed dye and one showing the green dye, with which we crafted the words UCL and iGEM as shown above.
3. Charactisation of Cambridge Part:BBa_K274003
E. coli turned dark green after transformation of this plasmid. This is unexpected as there is no promoter upstream of the operon.
Diagnostic DNA gel was run to test all 4 restriction sites. BBa_K274003 was cut with XbaI, PstI, EcoRI & SpeI, XbaI & PstI. A gel picture is shown as below.
This plasmid did not cut with Xba I restriction site in our hands. As such we could not use it to assemble after other parts.
Click on the following link for the [http://partsregistry.org/Part:BBa_K274003:Experience#User_Reviews Cambridge Part Review]


 "
"








 Twitter
Twitter Facebook
Facebook UCL
UCL Flickr
Flickr YouTube
YouTube