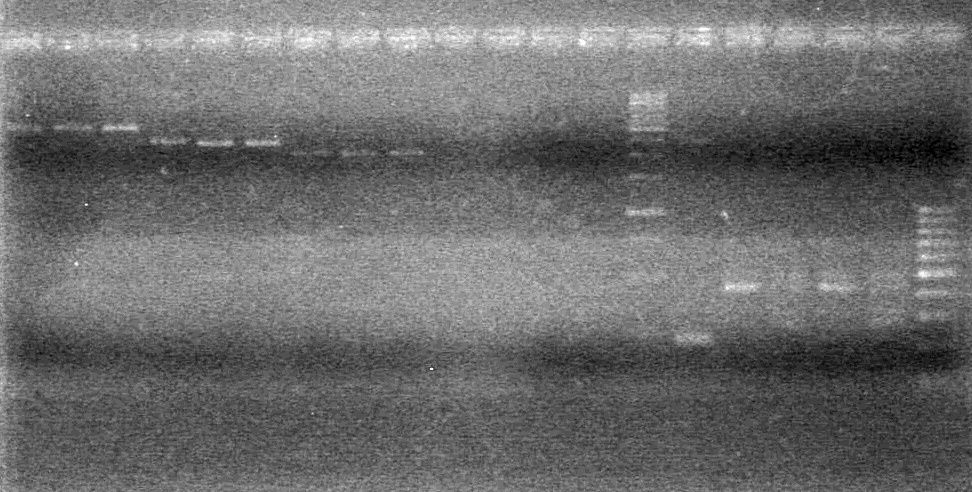
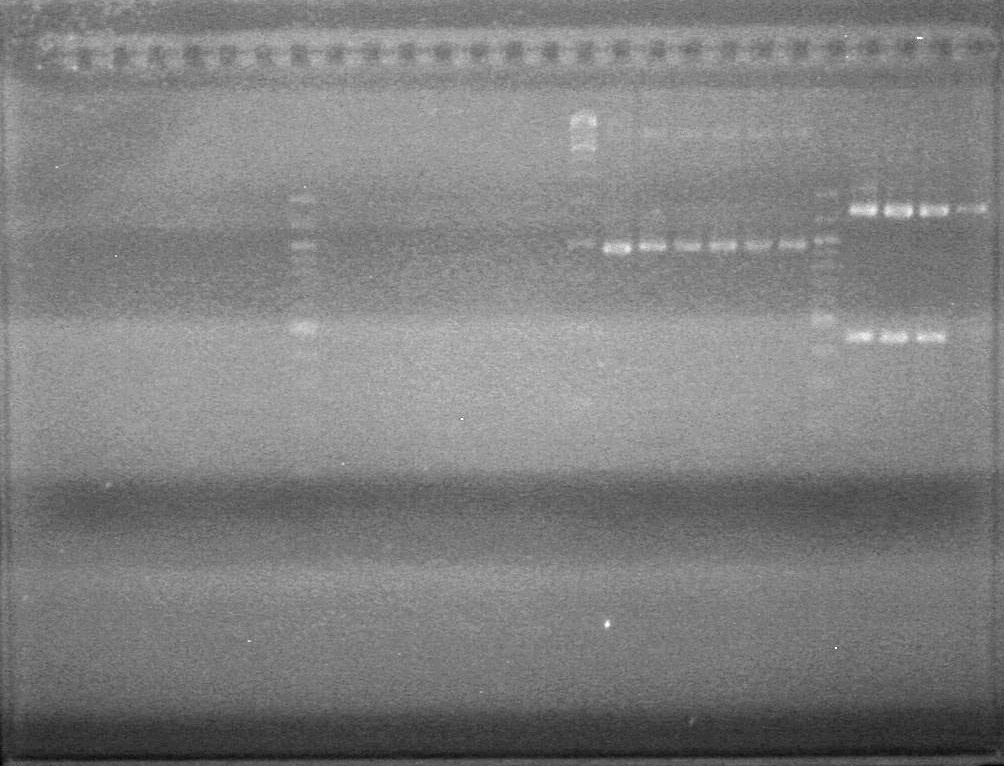
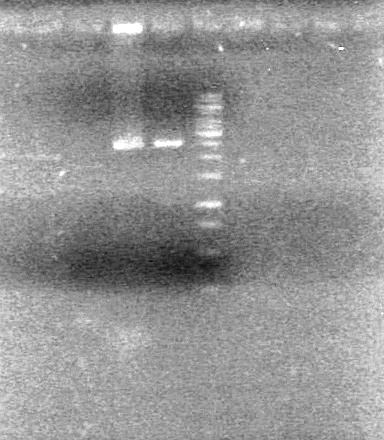
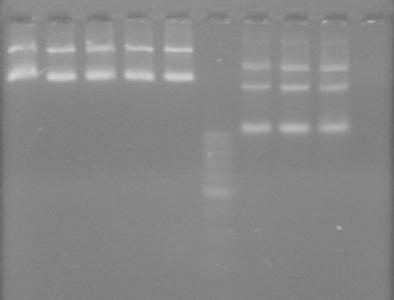
Used iGEM thermocycler setting: PFU.
| Lane | Sample | Volume
Sample (µL) | Volume Loading
Dye (µL) |
| 1 | xylE F-S 1 | 5 | 1 |
| 2 | xylE F-S 2 | 5 | 1 |
| 3 | xylE F-S 3 | 5 | 1 |
| 4 | xylE F-S 4 | 5 | 1 |
| 5 | xylE F-S 5 | 5 | 1 |
| 6 | xylE F-S 6 | 5 | 1 |
| 7 | 100bp Ladder (NEB) | 0.5 | 1(+5 H20) |
| 8 | xylE S-F 1 | 5 | 1 |
| 9 | xylE S-F 2 | 5 | 1 |
| 10 | xylE S-F 3 | 5 | 1 |
| 11 | xylE S-F 4 | 5 | 1 |
| 12 | xylE S-F 5 | 5 | 1 |
| 13 | xylE S-F 6 | 5 | 1 |
| 14 | Empty | | |
| 15 | PCR of K118021-B0015 (ADS) | 1 | 1 |
| 16 | Empty | | |
| 17 | Empty | | |
Results:
- Both KG PCR (Standard-Fusion and Fusion-Standard) amplified
- ADS PCR Amplified, but no insert present.
September 23, 2010
JV
Objective: Characterized catechol degradation by xylE enzyme
Method: Measured absorbance of catechol (275nm) and 2-hydroxymuconate semialdehyde (380nm).
- Protocol:
- 1) Grow cells in M9 minimal medium
- 2) Take 1/10 dilution of cells
- 3) Introduce 1µL of 0.05M catechol solution into the cell dilution. (Final concentration of 50µM;).
- 4) Quench the reaction with 5A% w/v trichloroacetate at certain time points. (0,15sec, 30sec, 45sec, 60sec, 2min, 3min, 4min, 5min, 10min).
- 5) Spin down cells.
- 6) Measure absorbance of supernatant.
Results: Cuvette used interfered with Spectra.
ADS
NOTE: In all transformations, heat shock step was missed. HOWEVER, all transformations showed significant number of colony forming units.
Objective: Move xylE (two biobrick; one with Fusion prefix, one with fusion suffix) into pSB1C3.
Method:
- Restriction of xylE PCR product and pSB1C3 (containing J04450 biobrick) via BioBrick Method using EcoRI-HF and PstI (Enzymes from NEB)
- Ligation of xylE PCR product and pSB1C3 via BioBrick Method using T4 DNA ligase (Enzyme from NEB)
- Transform into Subcloning Efficiency Compentent DH5α Cells (Invitrogen)
Results: TBD
Follow-up: TBD
Objective: Create glycerol stocks of <partinfo>J04450</partinfo> in pSB1A3 and pSB1T3 for use in RFP-BioBrick Assembly.
Method: Transform into Subcloning Efficiency Competent DH5α Cells (Invitrogen)
Obtained all plasmid DNA from 2010 Kit Plate 1
- J04450 in pSB1A3 - Well 1C
- J04450 in pSB1T3 - Well 7A
Results: Obtained TNTC colonies
Follow-up:
- Grow overnight cultures
- Generate Glycerol Stocks
- Generate Plasmid DNA via Maxiprep
Objective: Create glycerol stocks of received synthesized (Mr. Gene) signal peptides.
Method: Transform into Subcloning Efficiency Competent DH5α Cells (Invitrogen) plasmid DNA containing the following BioBricks:
- 1) <partinfo>K331007</partinfo> - β-lactamase Bla Signal Sequence
- 2) <partinfo>K331008</partinfo> - Outer Membrane Protein ompA
- 3) <partinfo>K331009</partinfo> - Heat Stable Toxin I
- 4) <partinfo>K331012</partinfo> - Penicillin Binding Protein DacA
- All inserts in pMA-T vector (Standard Mr. Gene vector)
Results: Obtained TNTC Cells
Follow-up:
- 1) Grow overnight cultures
- 2) Purify pDNA
- 3) Move into pSB1C3 plasmid
- 4) Verify sequence
- 5) Submit to registry for sequencing
TF, MC
Objective: Obtain a preparation of J04450 (RFP).
ethod: Used Plasmid DNA Purification by Alkaline Lysis (Large Scale AKA Maxiprep protocol.
September 24, 2010
ADS
Objective: Generate plasmid DNA of <partinfo>E1010</partinfo> for downstream PCR
Method: Transform plasmid DNA into Subcloning Efficiency Competent DH5α Cells (Invitrogen)
DNA obtained from 2010 Kit Plate 1 Well 18F (E1010 in pSB2K3)
Results: Obtained TBD colonies
Follow-up:
- Grow overnight cultures (Generate glycerol stocks)
- Purify plasmid DNA (Generate pDNA stocks)
- PCR to add terminal fusion standards
Objective: Create glycerol stocks of J04450 in pSB1K3 for use in RFP-BioBrick Assembly.
Method: Transform into Subcloning Efficiency Competent DH5α Cells (Invitrogen)
Obtained plasmid DNA from 2010 Kit Plate 1 well 5A (J04450 in pSB1K3)
Results: Obtained TNTC colonies
Follow-up:
- Grow overnight cultures
- Generate Glycerol Stocks
- Generate Plasmid DNA via Maxiprep
September 25, 2010
JV
Objective: Extract Plasmid DNA from DH5α cells.
Method:Qiagen spin column protocol.
- <partinfo>K331007</partinfo> (in pMA-T vector)
- <partinfo>K331008</partinfo> (in pMA-T vector)
- <partinfo>K331009</partinfo> (in pMA-T vector)
- <partinfo>K331012</partinfo> (in pMA-T vector)
Cells containing plasmids were put into glycerol stocks and put into HJ's -80oC.
ADS
Objective: Move plasmid DNA made by JV (above, Sept 25, 2010) into pSB1C3 for submission to the Standard Registry of Parts.
Method:
- 1) Digest at 37o for 10 min with PstI and EcoRI:
- pSB1C3 containing <partinfo>J04450</partinfo>
- pMA-T with <partinfo>K331007</partinfo>, <partinfo>K331008</partinfo>, <partinfo>K331009</partinfo>, and <partinfo>K331012</partinfo>.
- 2) Heat Kill at 80oC for 20 min
- 3) Mix 2µL of pSB1C3 with each biobrick, ligate with T4 DNA ligase for 10 min
- 4) Transform into subcloning efficiency DH5α cells, plate on ampicillin plates.
Results: Obtained RESULT!!! positive colonies
Objective: Assemble KG's C-terminal fusion xylE (<partinfo>K331021</partinfo>) (UNCONFIRMED) and C-terminal Arginine + dT (<partinfo>S04261</partinfo>) to create <partinfo>K331011</partinfo>.
Method:
- 1) Digest at 37o for 10 min:
- pSB1C3 containing <partinfo>J04450</partinfo> with PstI and EcoRI
- <partinfo>K331021</partinfo> with EcoRI, SpeI
- <partinfo>S04261</partinfo> with XbaI and PstI
- 2) Heat Kill at 80oC for 20 min
- 3) Mix 2µL of pSB1C3 with each biobrick, ligate with T4 DNA ligase for 10 min
- 4) Transform into subcloning efficiency DH5α cells, plate on ampicillin plates.
Results: Obtained RESULT!!! positive colonies
September 26, 2010
ADS
Objective: Purify plasmid DNA from 5mL cultures grown from transformations of KG's ligated PCRs containing (UNCONFIRMED) <partinfo>K331020</partinfo> and <partinfo>K331021</partinfo>.
Method: Used Qiagen spin column method with BioBasic EZ-10 spin columns.
Objective: Assemble the following:
- <partinfo>K331020</partinfo> + <partinfo>B0015</partinfo> to obtain To be named biobrick
- <partinfo>K331021</partinfo> + <partinfo>S04261</partinfo> to obtain To be named biobrick
Method:
- Digest <partinfo>K331020</partinfo> and <partinfo>K331021</partinfo> with EcoRI and SpeI
- Digest <partinfo>B0015</partinfo> and <partinfo>S04261</partinfo> with XbaI and PstI
- Digest <partinfo>J04450</partinfo> in pSB1C3 with EcoRI and PstI
Incubate each at 37oC for 10 min
Heat kill at 80oC for 20 min
Ligate with T4 Ligase for 10 min at room temperature
Transform into DH5&alpha subcloning efficiency cells
Results: Obtained RESULTS!!!! positive colonies.
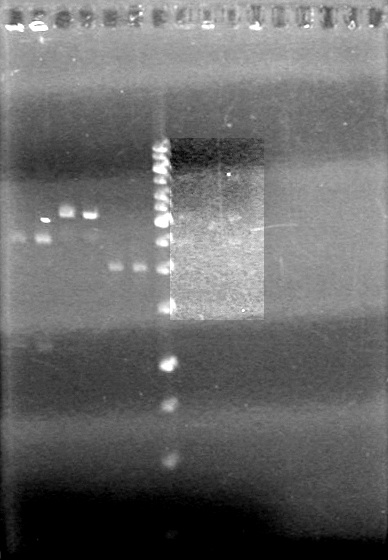
Also analyzed minipreps on 1.5% TAE Agarose Gel
GEL!!!
September 29, 2010
ADS
Received synthesized parts from Mr. GENE (all in pMA vectors)
- <partinfo>K331022</partinfo>
- <partinfo>K331023</partinfo>
- <partinfo>K331024</partinfo>
- <partinfo>K331025</partinfo>
Each tube contained 5µg pDNA; reconstitute in 50µL of TE buffer to get 100ng/µL concentration
Transform 1µL into DH5α subcloning efficiency cells
Obtained TNTC colonies
Started overnight 5mL cultures for miniprep and insertion into pSB1C3 vector.
AV
Objective: PCR the N-term fusion tag onto RFP (E1010) using sloppy annealing of the YFP/CFP fusion primers.
Composition of each PCR tube:
| Component | Volume(µL) |
| 10x Pfu Buffer w/ MgSO4 | 2 |
| dNTP (10mM) | 1 |
| N-term Fus Prefix | 1 |
| Biobrick Suffix | 1 |
| MilliQ H2O | 12.8 |
| Template DNA (J04450) | 2 |
| Pfu DNA Polymerase | 0.2 |
PCR conditions:
| Step | Temperature (oC) | Time (mins) | Number of cycles |
| Initial Denaturation | 95 | 2 | 1 |
| Denaturation | 95 | 0.5 | 25 |
| Annealing | 47.6, 50.5, 53.8 (gradient) | 0.5 | 25 |
| Extension | 72 | 2 | 25 |
| Final Extension | 72 | 10 | 1 |
Results: Gel has not been run. Have decided to order RFP specific primers for next year.
 "
"