Team:Imperial College London/Results/Exp3
From 2010.igem.org
(New page: {{:Team:Imperial_College_London/Templates/Header}} {{:Team:Imperial_College_London/Templates/ResultsHeader}}) |
|||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College_London/Templates/Header}} | {{:Team:Imperial_College_London/Templates/Header}} | ||
{{:Team:Imperial_College_London/Templates/ResultsHeader}} | {{:Team:Imperial_College_London/Templates/ResultsHeader}} | ||
| + | |||
| + | {| style="width:900px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Experiment 3 | Characterizing kinetic parameters of C2,3O in whole cells | ||
| + | |- | ||
| + | | | ||
| + | {|style="width:850px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | | | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:304px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Xyle reaction pic.jpg|300px]] | ||
| + | |- | ||
| + | |Catechol converted to 2-hydroxymuconic semialdehyde by C2,3O. | ||
| + | |} | ||
| + | </div> | ||
| + | | | ||
| + | In our Parasight project the XylE gene product, C2,3O enzyme, is the means by which an output signal is generated. If the parasite is present is the water supply, the bacterial system is activated by an input signal which produces the yellow color, the positive output signal. Test assays in the lab on colonies of cells expressing XylE gene, become yellow only seconds after addition of catechol (100mM). From this it was realized our group that the kinetics of the C2,3O enzyme are such that make it an attractive candidate as a reporter enzyme. So, we decided that further characterization of the kinetics of the C2,3O might help identify a really efficient reporter for research centers. | ||
| + | |} | ||
| + | |||
| + | {|style="width:850px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | | | ||
| + | In this experiment we investigated how the velocity of the reaction (catechol converted to 2-hydroxymuconic semialdehyde by C2,3O) varies with increasing initial concentrations of catechol, the substrate. Overnight cultures of XylE transformed E.coli were plated on a 96 wells plate at an optical density (O.D.) of 0.5 units of absorbance. (The optimum initial O.D. of cells was determined in a previous experiment. This O.D. may vary across different labs as the sensitivity of the spectrophotometer/plate reader is a factor). Catechol concentrations ranging from 0.1-1mM were made from 100mM stock solution and added to the cells of the 96-well plate. The velocity of the reaction was monitored using the plate reader spectrophotometer as the reaction is directly proportional to the production of yellow color. The yellow output was measured at 380nm wavelength, while cell density at 600nm. The XylE gene expression was under the influence of J23101 promoter. | ||
| + | | | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:304px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:IC_Assay_3_sept.jpg|300px]] | ||
| + | |- | ||
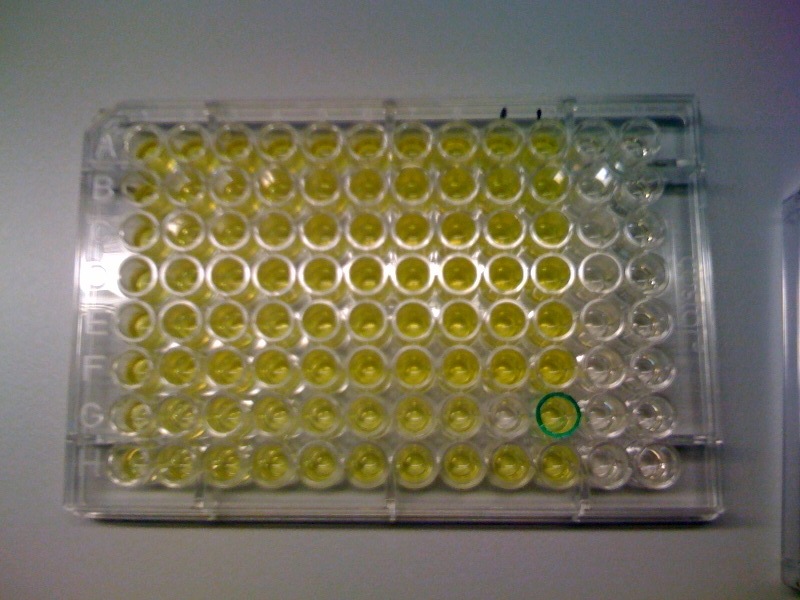
| + | |Catechol assay on XylE-trasformed cells in a 96-well plate (A to H decreasing cell concentration, 1-10 decreasing catechol concentration, column 11 and 12 negative and control). | ||
| + | |} | ||
| + | </div> | ||
| + | |} | ||
| + | |||
| + | {|style="width:850px;background:#f5f5f5;text-align:justify;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | | | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:304px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Catechol assay graph (0.2-1).jpg|300px]] | ||
| + | |- | ||
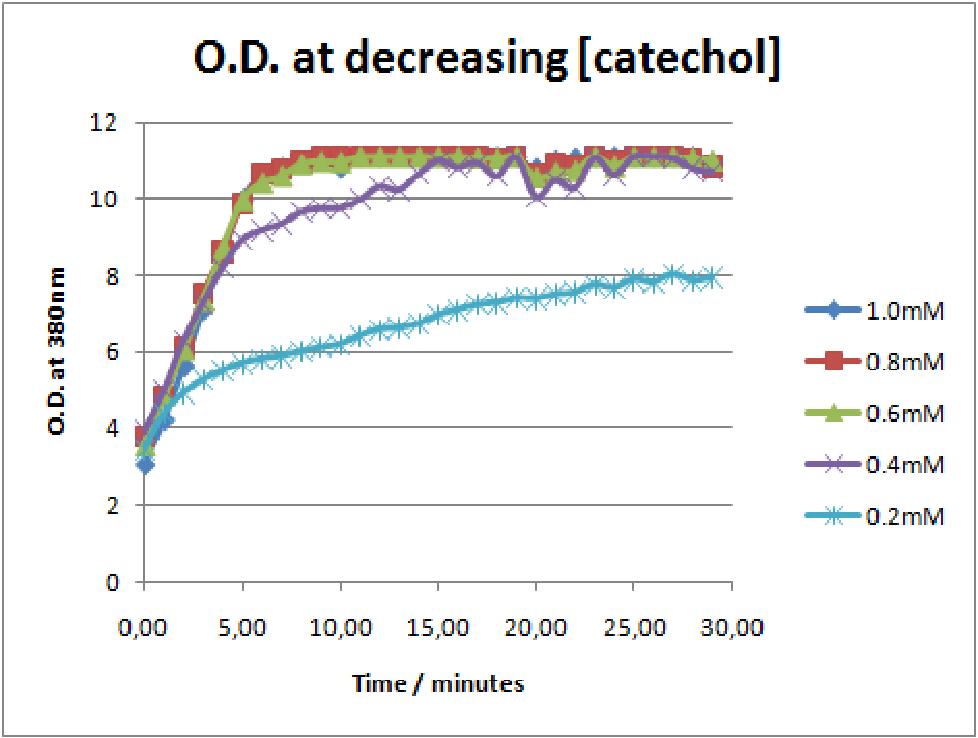
| + | |The first catechol assay data analysis. The assay involved addition of catechol concentrations of 0.2, 0.4, 0.6, 0.8 and 1mM to 0.5 O.D. of XylE transformed E.coli cell cultures. The XylE gene was under the influence of J23101 promoter. | ||
| + | |} | ||
| + | </div> | ||
| + | | | ||
| + | In this experiment we investigated how the velocity of the reaction (catechol converted to 2-hydroxymuconic semialdehyde by C2,3O) varies with increasing initial concentrations of catechol, the substrate. Overnight cultures of XylE transformed E.coli were plated on a 96 wells plate at an optical density (O.D.) of 0.5 units of absorbance. (The optimum initial O.D. of cells was determined in a previous experiment. This O.D. may vary across different labs as the sensitivity of the spectrophotometer/plate reader is a factor). Catechol concentrations ranging from 0.1-1mM were made from 100mM stock solution and added to the cells of the 96-well plate. The velocity of the reaction was monitored using the plate reader spectrophotometer as the reaction is directly proportional to the production of yellow color. The yellow output was measured at 380nm wavelength, while cell density at 600nm. The XylE gene expression was under the influence of J23101 promoter. | ||
| + | |} | ||
| + | |||
| + | The results of the experiment confirmed previous hints of the huge catalytic turnover rate of the C(2,3)0 enzyme. The graph on the left illustrates production of yellow product per min. The slope at the initial stages of the reaction (first minutes) would represent the rate of the reaction, that is the initial Velocity. By calculating initial velocities of the reaction at different substrate concentrations, one can plot a Michaelis-Menten curve to extract kinetic parameters such as Vmax, Km and catalytic turnover number. | ||
| + | |||
| + | Several attempts were made to determine these parameters on whole cells. Unfortunately due to the actual nature of the enzyme, velocity of reaction and technical limitations of lab equipement this has been proven impossible. To start with one of the limitation was that the color output and the extinction coefficient of the yellow product is so large, that the system saturates within the first minutes after catechol addition to cell cultures. The spectrophotometer and plate reader reaches its detection maximum reading value from the start of the reaction. Also due to the high velocity of the reaction, in the time frame of about 10 seconds that are needed to load the plate into the plate reader after catechol addition, the reaction has already proceeded too far. So initial velocity measurements of the reactions needed for construction of Michaelis-Menten curve are inaccurate. These problem could be overcome by decreasing total enzyme concentration. In whole cell cultures this would be possible by decreasing cell density in each well as each cell contain an average C2,3O enzyme concentration. However, previous experiments showed that cell cultures with optical densities lower than 0.5 O.D. introduce serious perturbations in the readings of the spectrophotometer and plate reader detectors. | ||
| + | |||
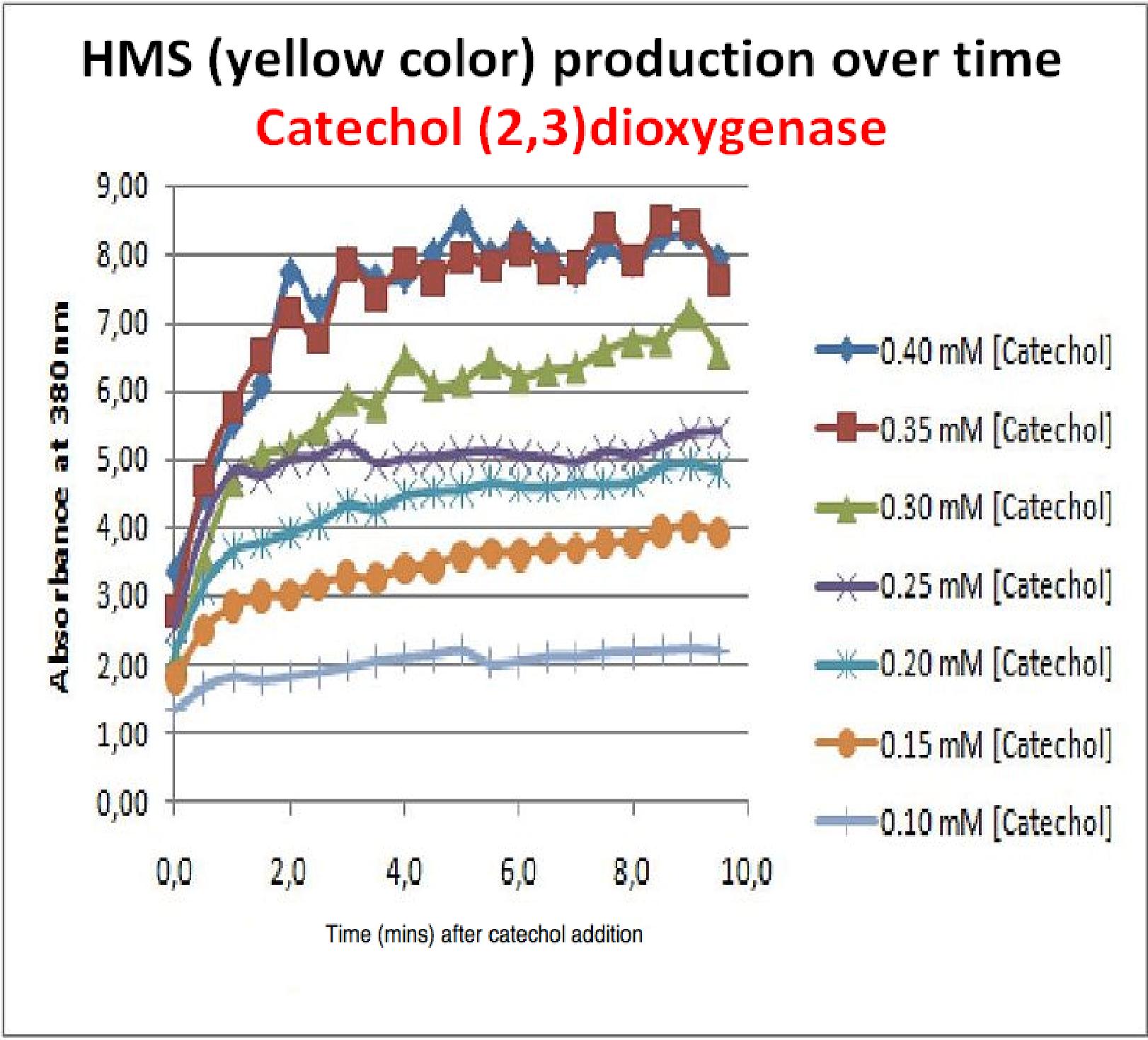
| + | Despite these limitations some useful data were extracted out of these assays. More specifically we acquired data that delineate the course of the reaction in terms of yellow product production over time at various catechol concentrations. The results of one of these assays is presented in the figure below. In order to extract data that will allow characterization of the kinetic parameters of catechol dioxygenase enzyme our lab team proposes purification of the protein from cell lysate, '''several fold dilution''', and in vitro characterization. | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:304px;background:#e7e7e7;text-align:center;font-family: helvetica, arial, sans-serif;color:#555555;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:HMS prod. over time curve for xylE.jpg|400px]] | ||
| + | |- | ||
| + | |Graph shows production of HMS (yellow product) over time after catechol addition at time 0 minutes. Different curves represent different catechol concentration added to the cell cultures. | ||
| + | |} | ||
| + | </div> | ||
| + | |} | ||
Revision as of 15:10, 27 October 2010
| Experimental Results | Exp 1 | Exp 2 | Exp 3 | Exp 4 | Exp 5 | Exp 6 | Exp 7 |
| Testing is a fundamental stage of the engineering design cycle and is a crucial part of charactrising BioBrick Standard Biological Parts so that other people can benefit from our work. We've compiled all our results on this page, detailing how the experiments were carried out and the significance of the data. | |
| Experiment 3 | Characterizing kinetic parameters of C2,3O in whole cells | ||||||
The results of the experiment confirmed previous hints of the huge catalytic turnover rate of the C(2,3)0 enzyme. The graph on the left illustrates production of yellow product per min. The slope at the initial stages of the reaction (first minutes) would represent the rate of the reaction, that is the initial Velocity. By calculating initial velocities of the reaction at different substrate concentrations, one can plot a Michaelis-Menten curve to extract kinetic parameters such as Vmax, Km and catalytic turnover number. Several attempts were made to determine these parameters on whole cells. Unfortunately due to the actual nature of the enzyme, velocity of reaction and technical limitations of lab equipement this has been proven impossible. To start with one of the limitation was that the color output and the extinction coefficient of the yellow product is so large, that the system saturates within the first minutes after catechol addition to cell cultures. The spectrophotometer and plate reader reaches its detection maximum reading value from the start of the reaction. Also due to the high velocity of the reaction, in the time frame of about 10 seconds that are needed to load the plate into the plate reader after catechol addition, the reaction has already proceeded too far. So initial velocity measurements of the reactions needed for construction of Michaelis-Menten curve are inaccurate. These problem could be overcome by decreasing total enzyme concentration. In whole cell cultures this would be possible by decreasing cell density in each well as each cell contain an average C2,3O enzyme concentration. However, previous experiments showed that cell cultures with optical densities lower than 0.5 O.D. introduce serious perturbations in the readings of the spectrophotometer and plate reader detectors. Despite these limitations some useful data were extracted out of these assays. More specifically we acquired data that delineate the course of the reaction in terms of yellow product production over time at various catechol concentrations. The results of one of these assays is presented in the figure below. In order to extract data that will allow characterization of the kinetic parameters of catechol dioxygenase enzyme our lab team proposes purification of the protein from cell lysate, several fold dilution, and in vitro characterization. |
 "
"