Output Amplification Model
Goals:
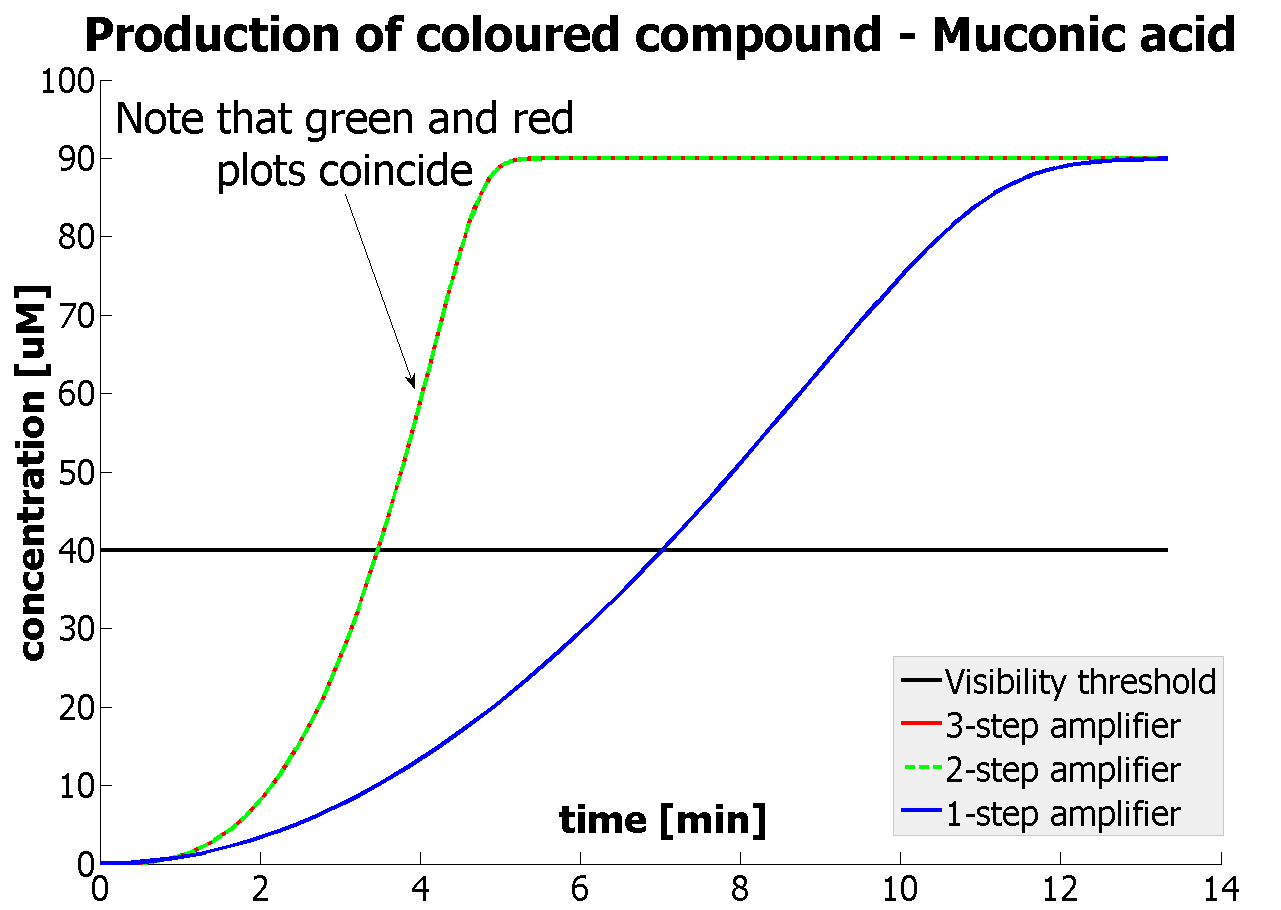
This model was mainly developed in order to determine whether simple production is better than 1-, 2- or 3-step amplification. Furthermore, an estimation of the speed of the response was desirable.
Elements of the system:
- Dioxygenase (blue on the diagrams below) is an enzyme that acts on catechol to produce a yellow output. In most of our models dioxygenase was treated as an output because it was found that active dioxygenase acting on catechol produces the coloured output within a split second.
- GFP-Dioxygenase fusion protein (GFP is shown green on the diagrams). Dioxygenase joined by the linker to GFP was assumed to be inactive.
- TEV protease (pink on the diagrams below) has the ability to cleave the GFP-Dioxygenase fusion protein, hence, it activates dioxygenase
- Split TEV protease (purple on the diagrams below) is an inactive split form of TEV mounted on coiled coils. It can be activated again by coiled coils being cleaved by another active TEV.
|
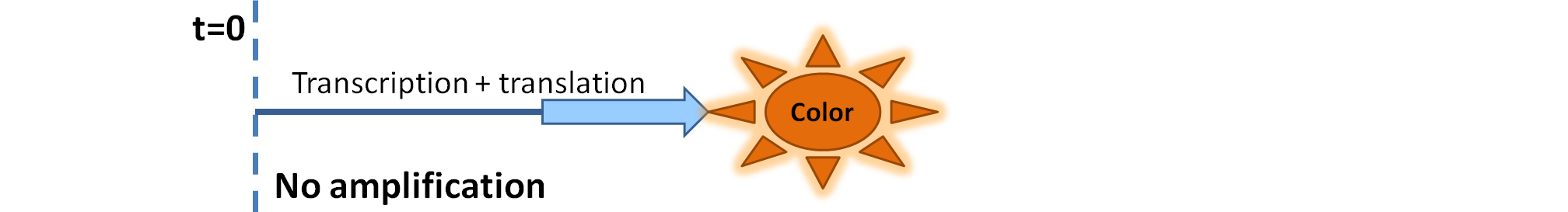
| Simple production upon activation of arbitrary colour output by transcription and translation indicated by the blue arrow.
|
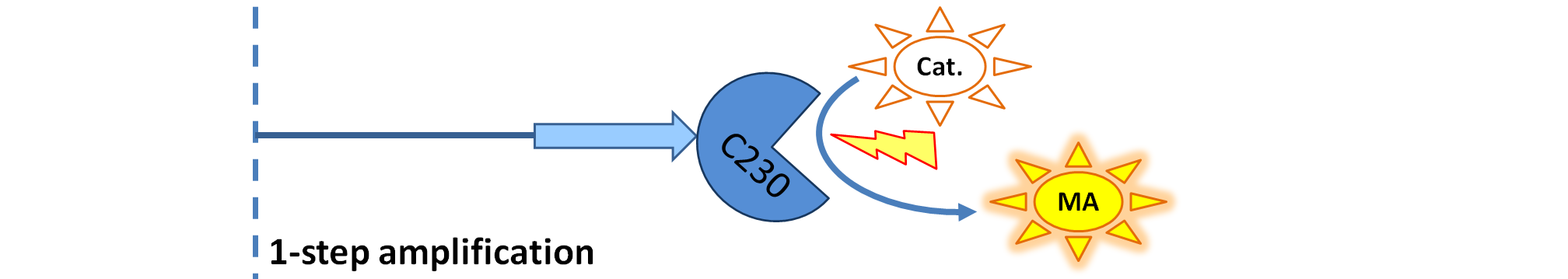
|
| Dioxygenase (C230) is simply produced. Upon activation at time t=0, it acts on catechol (cat.) to produce yellow output - muconic acid. Catechol is not shown to be produced by cell as it is added by person at arbitrary time.
|
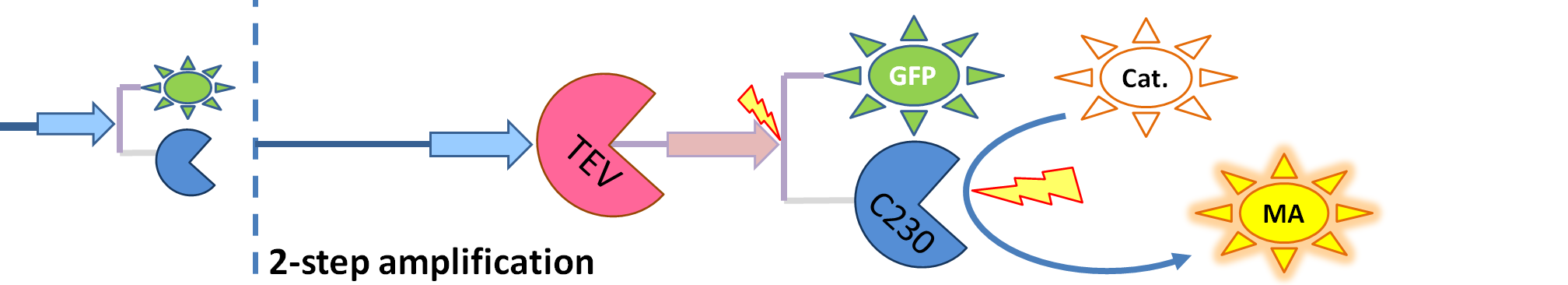
|
| The species that are shown in front of vertical line which indicates beginning of experiment mean that they have been accumulated beforehand in the cell. TEV protease activates inactive dioxygenase which acts on catechol to produce colour.
|
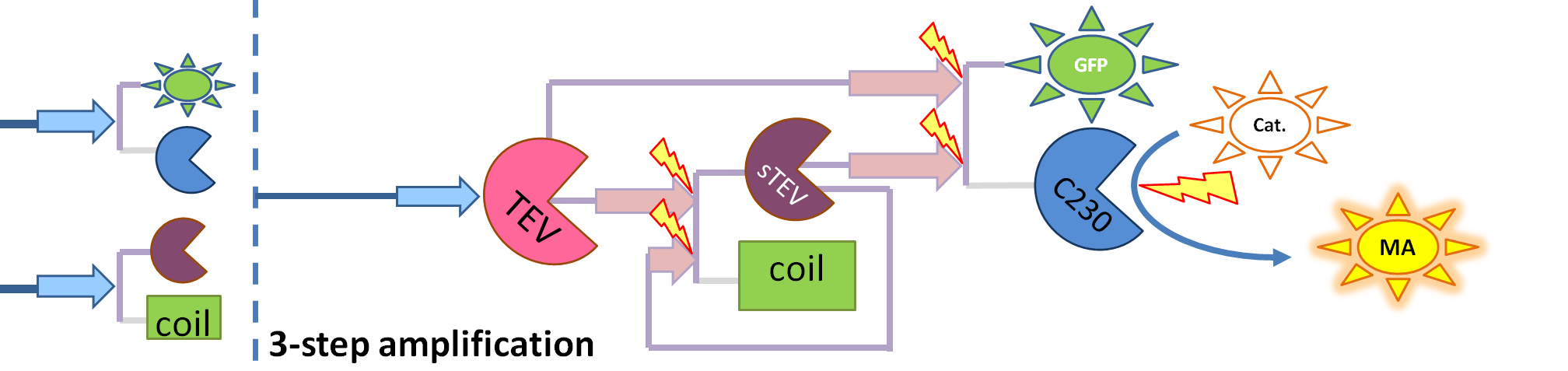
|
| This diagram introduces inactive split TEV protease attached to a coiled-coil as the third amplification step. Both inactive compounds have active site for TEV to activate tehm which results in multiple possibilities of action.
|
Major assumptions:
- The chemical and enzymatic reactions are modelled according to the Law of Mass Action.
- Our model assumes that the modelled system is inert within the bacterial body or that reactions with other species within the bacterium is negligible. For example, the TEV protease is not supposed to cleave other molecules due to its specifity.
Signalling Modelling Model
Goals:
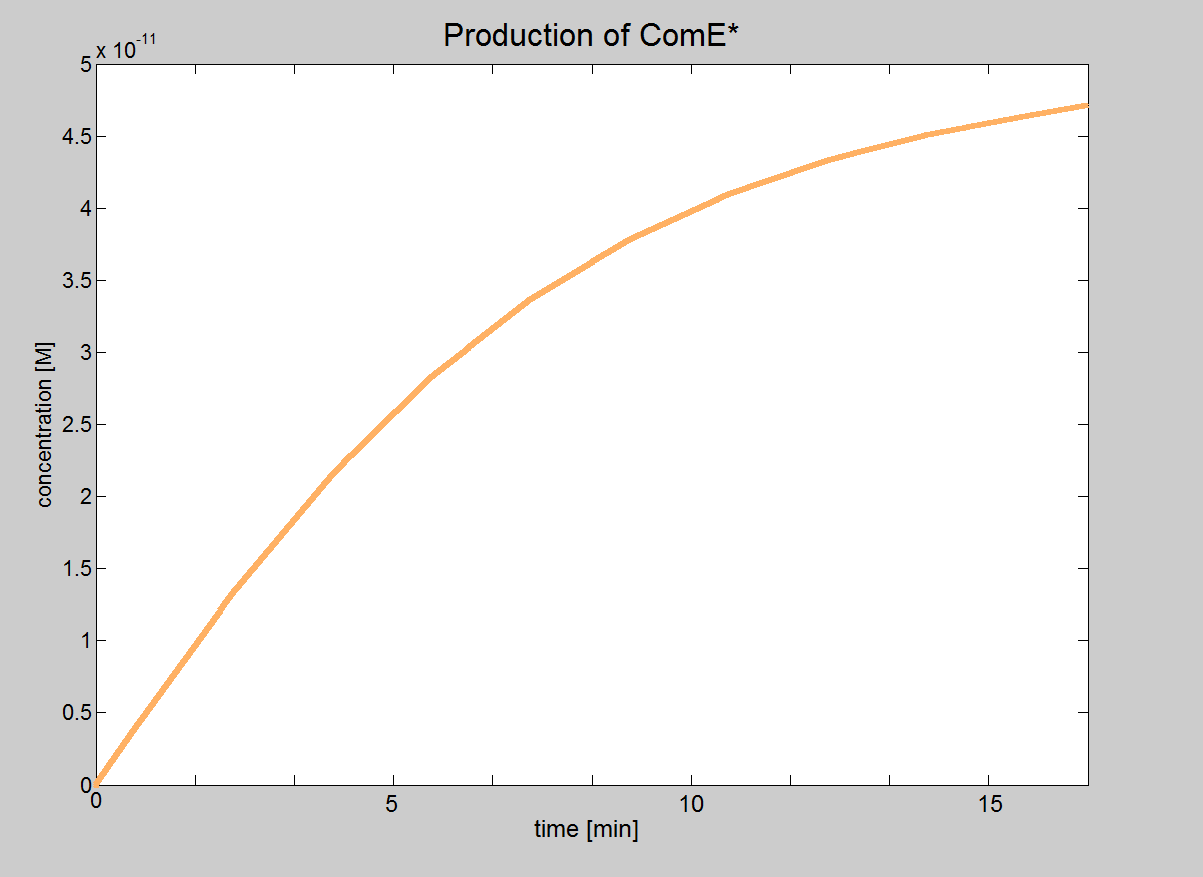
The aim of this model is to determine under which conditions the signalling transduction will happen in our bacteria.
Elements of the system:
- The signalling model consists of ComD and ComE which are expressed by our bacteria, as well as AIP and Phosphate. These species are all assumed to be present in the cell at a sufficient concentration.
- The ComD receptor is activated by the AIP. This triggeres phosphorylation of the ComD receptor. The Phosphate group of the ComD receptor then binds to ComE. The phosphorylated ComE binds to the DNA and acts as a transcription factor.
Major assumptions:
- ComD and ComE are present in the cell/cell wall at a high concentration. ComD and ComE are both in steady-state, so the production and degradation constants are negligible.
- AIP and Phosphate are present inside/outside the cell at a high concentration. The degradation rates for these two species are negligible.
- Phosphorylation of the ComD receptor is modelled as an enzymatic reaction, neglecting the formation of an intermediate complex.
Surface Protein Model
Goals:
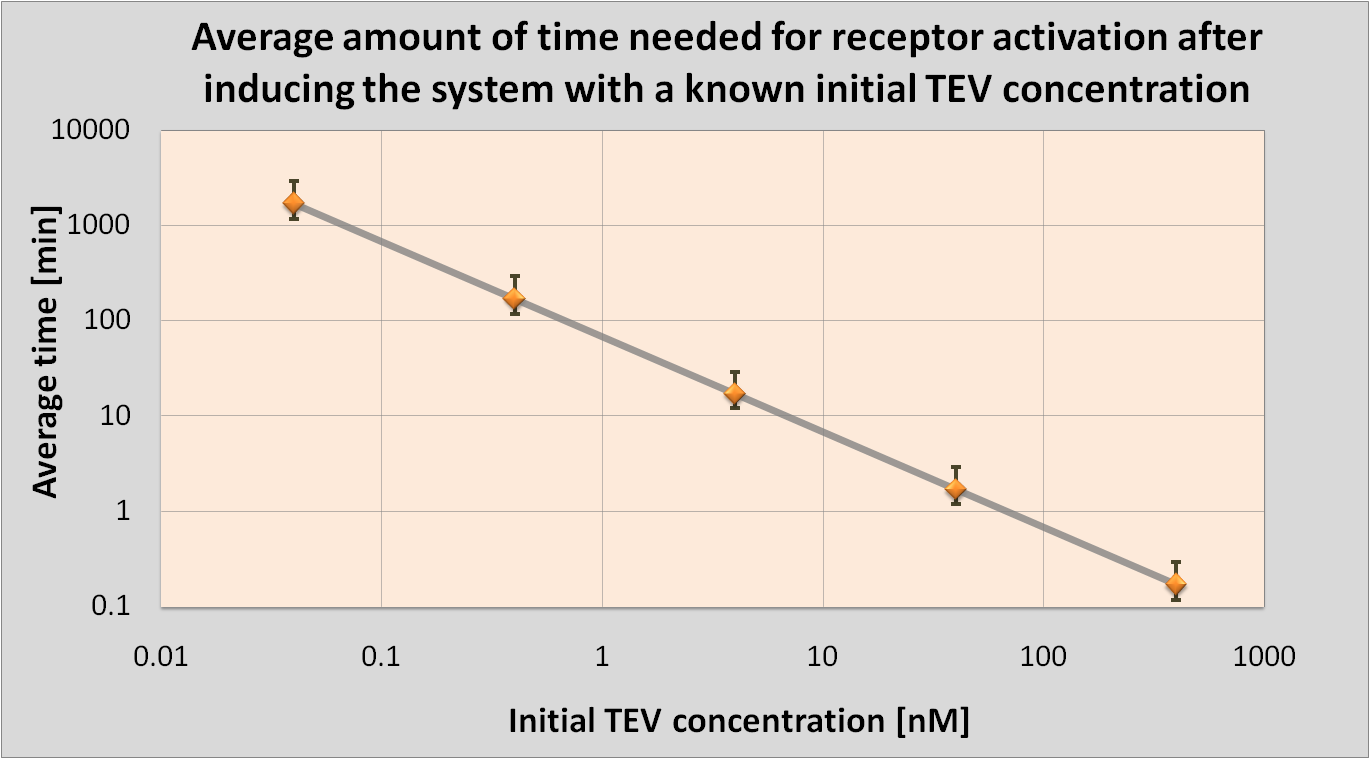
The aim of this model is to determine the concentration of Schistosoma elastase or TEV protease that should be added to the bacteria in order to trigger a response. This would allow us to correlate the required concentration for the activation with the concentration of Schistosoma elastase in the lake.
It was also attempted to model how long it takes for the protease or elastase to cleave the required amount of peptides.
Elements of the system:
- The surface protein consists of a cell wall binding domain, linker, AIP (Auto Inducing Peptide)
- Schistosoma elastase (this is the enzyme released by the parasite) cleaves AIP from the cell wall binding domain at the linker site. In the laboratory we used TEV protease as we could not obtain the Schistosoma elastase.
- The ComD receptor is activated (i.e. AIP concentration is high enough).
Major assumptions:
- The chemical and enzymatic reactions are modelled according to the Law of Mass Action.
- Our model assumes that the modelled system is inert within the bacterial body or that reactions with other species within bacterium is negligible. For example, the TEV protease is not supposed to cleave other molecules due to its specifity.
- Due to our carefully chosen cell concentrations, the diffusion of free AIPs could be neglected. However, this restricts the model to the considered cell concentrations only.
- The threshold for receptor activation was defined by one specific value as opposed to considering intermediate states between fully "off" and "on".
|
 "
"