Team:Freiburg Bioware/NoteBook/Labjournal/September2
From 2010.igem.org
(→Continuation with Colony-PCR of pSB1C3_CD and ViralBrick motif Z34C) |
(→Testtransduction of the final modified capsid coding constructs) |
||
| Line 170: | Line 170: | ||
Aim of the experiment: | Aim of the experiment: | ||
| + | <br/> | ||
At the beginning of the project 22 silent nucleotide exchange mutations were introduced theoretically into the capsid coding construct to make it compatible with the RFC standards and to have single cutting restriction enzymes flanking the 453 and the 587 loop sequence. During the experiments two point mutations had to be dismissed because either a first test transduction showed that the construct was no more working or the insertion of the synthesized gene posed serious problems because the restriction enzyme did not work.<br> | At the beginning of the project 22 silent nucleotide exchange mutations were introduced theoretically into the capsid coding construct to make it compatible with the RFC standards and to have single cutting restriction enzymes flanking the 453 and the 587 loop sequence. During the experiments two point mutations had to be dismissed because either a first test transduction showed that the construct was no more working or the insertion of the synthesized gene posed serious problems because the restriction enzyme did not work.<br> | ||
| - | '''Finally we have a construct that is shown to produce infectious particles comparable to the current AAV systems and carriey 20 point mutations. Luckily the Adeno-associated Virus is compatible to the RFC standard and the idea to replace the loop sequences via ViralBricks works! ''' | + | '''Finally we have a construct that is shown to produce infectious particles comparable to the current AAV systems and carriey 20 point mutations. Luckily the Adeno-associated Virus is compatible to the RFC standard and the idea to replace the loop sequences via ViralBricks works! '''<br /> |
| - | + | <br /> | |
Impression from the moment: | Impression from the moment: | ||
| - | [[Image:Freiburg10_Sieg auf ganzer Linie.jpg|thumb| | + | <br /> |
| - | + | [[Image:Freiburg10_Sieg auf ganzer Linie.jpg|thumb|center|700px|The construct with the insertion of the Rep and the Cap synthesis worked! Reintroduction of the KpnI restriction site recovered the functionality of the viral genome!]] | |
| - | + | ||
| - | + | ||
====<p style="font-size:15px; background-color:#66bbFF;"><b>Miniprep of serveral constructs</b></p>==== | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Miniprep of serveral constructs</b></p>==== | ||
Revision as of 14:29, 19 September 2010

Contents |
September
124. labday 19.09.2010
Continuation with Colony-PCR of pSB1C3_CD and ViralBrick motif Z34C
Investigator: Bea
Comment: Since the colony PCR of the last try did not work exactly the way we expected it, another colony PCR approach with new primers was conducted.
Loading plan:
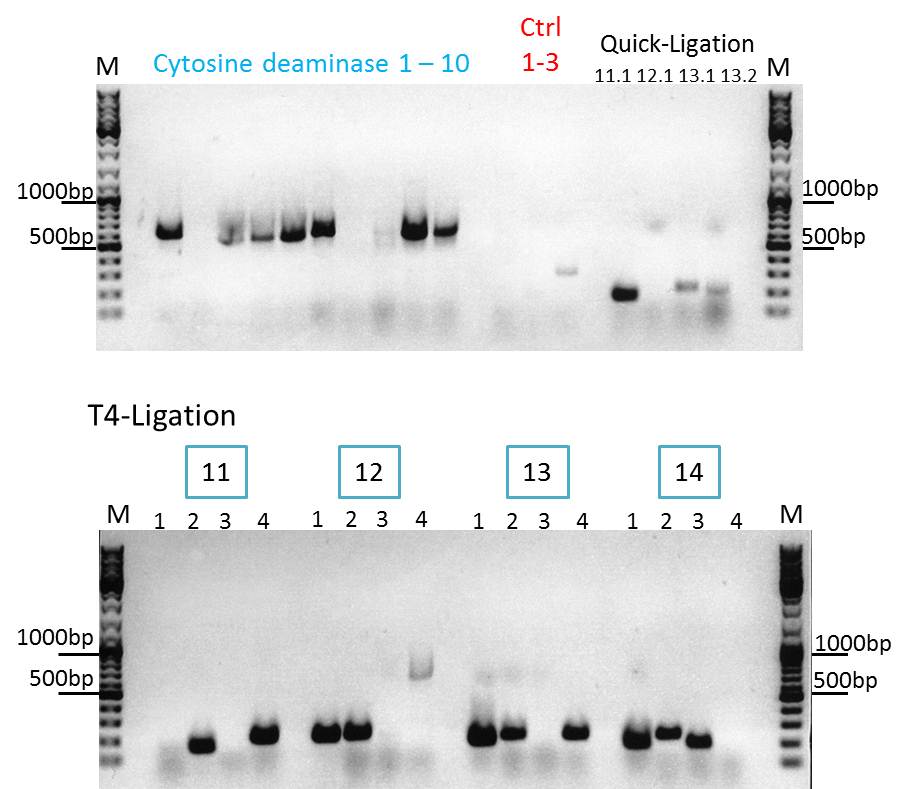
upper lanes
M CD1 CD2 CD3 CD4 CD5 CD6 CD7 CD8 CD9 CD10 +ctrl(CFP) +ctrl(BAP) -ctrl(pAAV_MCS) 11Q1 12Q1 13Q1 13Q2 M
lower lanes
M 11T41 11T42 11T43 11T44 12T41 12T42 12T43 12T44 13T41 13T42 13T43 13T44 14T41 14T42 14T43 14T44 M
Expected sizes of the PCR products are:
- Cytosine deaminase (CD) = 1300bp
- Z34C loops insertion motif = 220bp
- Positive control 1 = CFP = 750bp
- Positive control 2 = BAP_587 = 170bp
Results:
The Colony PCR of the Cytosine Deaminase (CD) did not work out. The detectable bands at around 700 bp correspond to CFP. Therefore, BioBrick production of the CD must be repeated. The following three bands represent the controls. Three different controls were used. The first corresponds to pSB1C3_CFP, the second to BAP_587 and third control is the negative control. Unfortunately now bands can be detected in the postive controls. A faint band can be seen in the negative control which is the only lane in which no band should be detectable.
The following bands after the three controls correspond to the approaches of the Z34C Viralbrick production. Some bands in the lower and the upper lanes show positive results. The corresponding inoculated overnight culture were centrifuged and prepared in order to perform a Mini-Prep tomorrow.
Testtransduction of the final modified capsid coding constructs
Investigator: Adrian
Aim of the experiment:
At the beginning of the project 22 silent nucleotide exchange mutations were introduced theoretically into the capsid coding construct to make it compatible with the RFC standards and to have single cutting restriction enzymes flanking the 453 and the 587 loop sequence. During the experiments two point mutations had to be dismissed because either a first test transduction showed that the construct was no more working or the insertion of the synthesized gene posed serious problems because the restriction enzyme did not work.
Finally we have a construct that is shown to produce infectious particles comparable to the current AAV systems and carriey 20 point mutations. Luckily the Adeno-associated Virus is compatible to the RFC standard and the idea to replace the loop sequences via ViralBricks works!
Impression from the moment:
Miniprep of serveral constructs
Investigator: Stefan
Glycerol stocks were prepared:
- B430 = pCerulean_ZEGFR:1907_Longlinker_VP2/3
- B431 = pCerulean_CFP_Middlelinker_VP2/3_insCap
- B432 = pCerulean_ZEGFR:1907_Shortlinker_VP2/3_insCap
Mini-Prep was performed according to the standard protocol
- P527 = pCerulean_ZEGFR:1907_Longlinker_VP2/3 c = 230,61 ng/µl
- P528 = pCerulean_CFP_Middlelinker_VP2/3_insCap c = 290,81 ng/µl
- P529 = pCerulean_ZEGFR:1907_Shortlinker_VP2/3_insCap c = 269,39 ng/µl
Several other constructs were preped, but need to be confirmed before being added to plasmid/glycerol stock.
 "
"