Team:Cambridge/Bioluminescence/Vibrio Characterisation
From 2010.igem.org
(→Arabinose to light) |
|||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Cambridge/Templates/headerMinimalprototype}} | {{:Team:Cambridge/Templates/headerMinimalprototype}} | ||
| - | {{:Team:Cambridge/Templates/headerbar|colour=# | + | {{:Team:Cambridge/Templates/headerbar|colour=#386abc|title=Project Vibrio: Characterisation}} |
This page describes characterisation for part [http://partsregistry.org/Part:BBa_K325909 BBa K325909], the [https://2010.igem.org/Team:Cambridge/Bioluminescence/Bacterial_Luciferases ''lux operon'' from ''Vibrio fischeri'']. | This page describes characterisation for part [http://partsregistry.org/Part:BBa_K325909 BBa K325909], the [https://2010.igem.org/Team:Cambridge/Bioluminescence/Bacterial_Luciferases ''lux operon'' from ''Vibrio fischeri'']. | ||
| Line 12: | Line 12: | ||
=Description= | =Description= | ||
{{:Team:Cambridge/Templates/RightImage|image=Cambridge-low.jpg|caption=E.Coli (Invitrogen TOP 10) cells transformed with [http://partsregistry.org/Part:BBa_K325909 BBa K325909] (blue light bulb) and [http://partsregistry.org/Part:BBa_K325219 BBa 325219] (red light bulb)}} | {{:Team:Cambridge/Templates/RightImage|image=Cambridge-low.jpg|caption=E.Coli (Invitrogen TOP 10) cells transformed with [http://partsregistry.org/Part:BBa_K325909 BBa K325909] (blue light bulb) and [http://partsregistry.org/Part:BBa_K325219 BBa 325219] (red light bulb)}} | ||
| - | This | + | This page described the lux operon from Vibrio fischeri. To relieve LuxR control we placed Lux C, D, A, B, E under the pBad promoter. |
| + | |||
| + | |||
| + | |||
| + | [[Image:250px-Cambridge-iGEMpixels.jpg|thumb|569px|center|'''Figure 1 - E.coli cells (Invitrogen TOP 10) transformed with [http://partsregistry.org/Part:BBa_K325909 BBa K325909] in a 96 well plate. ''']] | ||
=Arabinose to light= | =Arabinose to light= | ||
| Line 19: | Line 23: | ||
{{:Team:Cambridge/Templates/Nolineheader|header=Data}} | {{:Team:Cambridge/Templates/Nolineheader|header=Data}} | ||
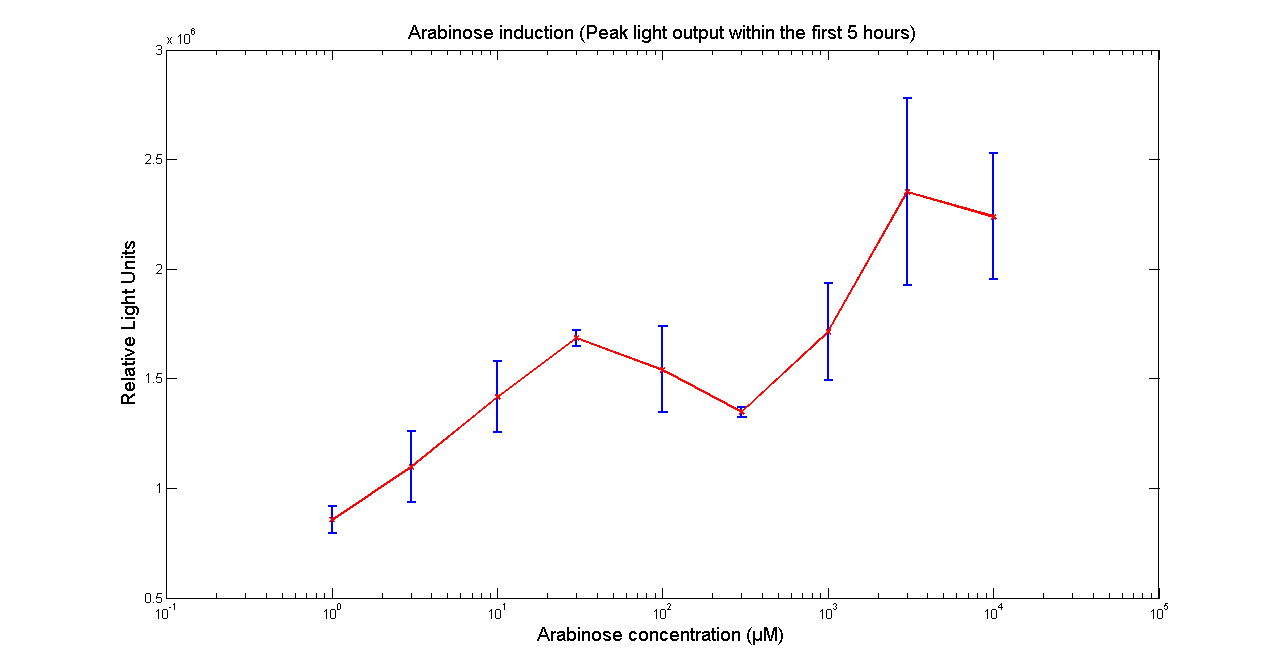
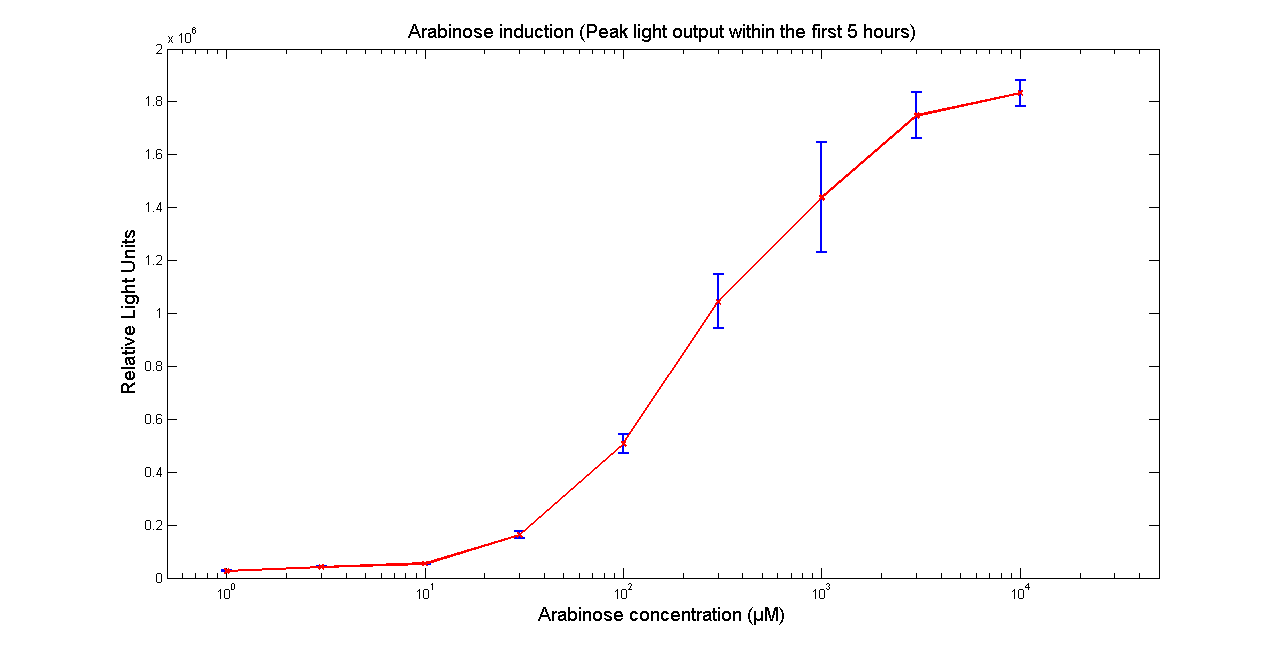
| - | [[Image:BBa_K325909Aracurve.png|thumb|569px|center|'''Figure 1 - Light output of <partinfo> | + | [[Image:BBa_K325909Aracurve.png|thumb|569px|center|'''Figure 1 - Light output of <partinfo>K325909</partinfo> as a function of Arabinose concentration in the media. The values correspond to the peak intensity within 5 hours of adding Arabinose to the media. Data points and error bars correspond to the mean and the standard deviation of 3 time repeats. ''']] |
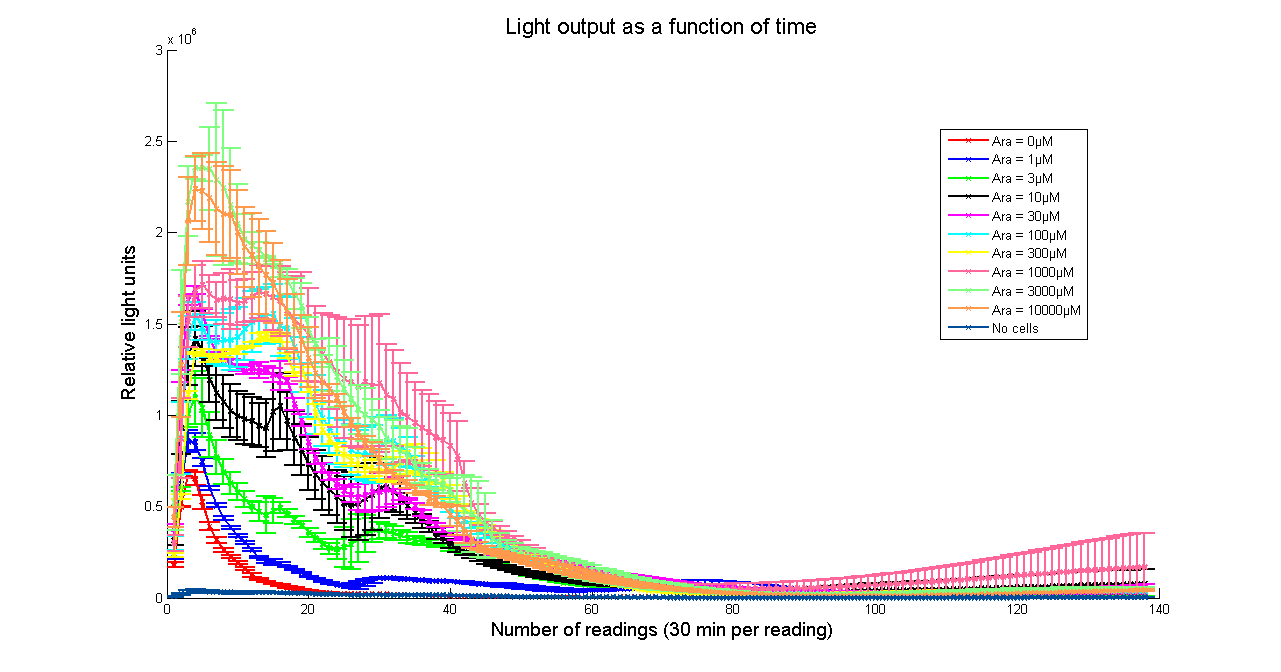
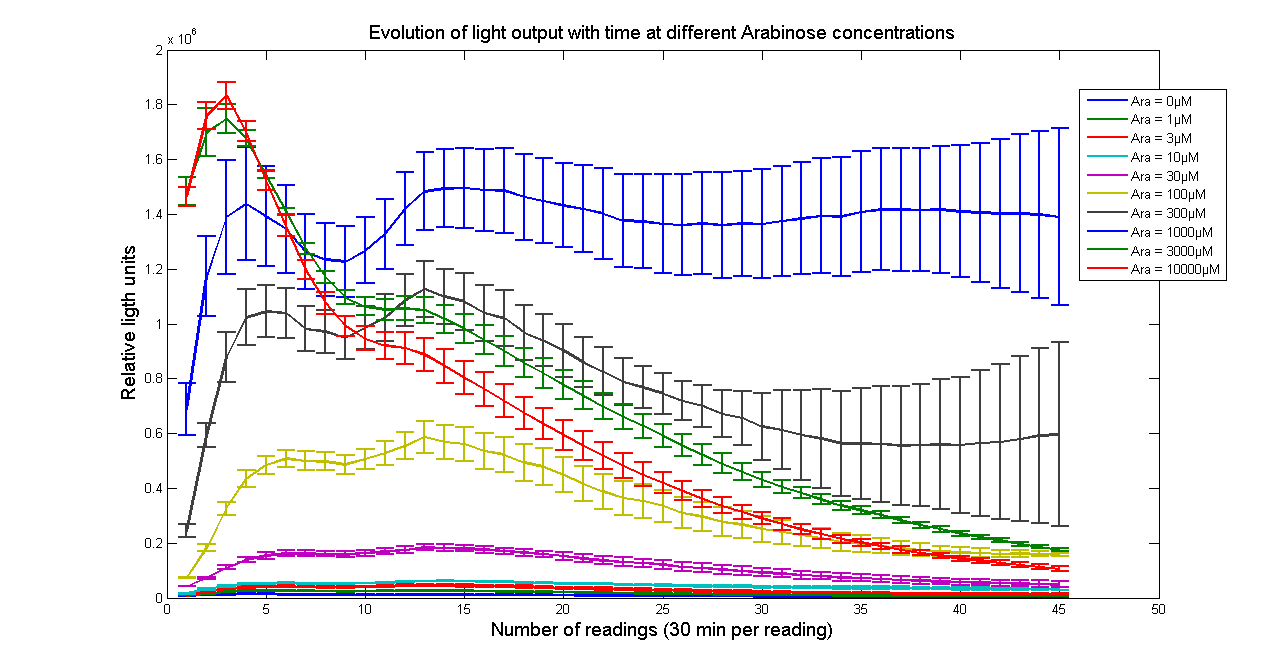
| - | [[Image:BBa_K325909timecourse.png|thumb|569px|center|'''Figure 2 - Evolution of luminescence with time at different Arabinose concentrations for <partinfo> | + | [[Image:BBa_K325909timecourse.png|thumb|569px|center|'''Figure 2 - Evolution of luminescence with time at different Arabinose concentrations for <partinfo>K325909</partinfo>. Measurements were taken every 30 minutes. Data points and error bars correspond to the mean and standard deviation of 3 time repeats. ''']] |
<center> | <center> | ||
{|{{Table}} | {|{{Table}} | ||
| Line 32: | Line 36: | ||
|} | |} | ||
</center> | </center> | ||
| - | |||
| - | |||
| - | |||
=H-NS mutants= | =H-NS mutants= | ||
| - | + | It has been shown that the expression of the Vibrio fischeri lux operon when cloned into E. coli was repressed. This repression was linked to the nucleoid protein H-NS. To investigate this effect we cloned the operon into mutant E.coli cells in which the expression of the H-NS protein had been modified. We used a [http://www.bmglabtech.com/products/microplate-reader/instruments.cfm?product_id=2 FLUOstar OPTIMA] microplate reader to quantify the light output. Protocols and plate reader settings used are given below. | |
{{:Team:Cambridge/Templates/Nolineheader|header=Data}} | {{:Team:Cambridge/Templates/Nolineheader|header=Data}} | ||
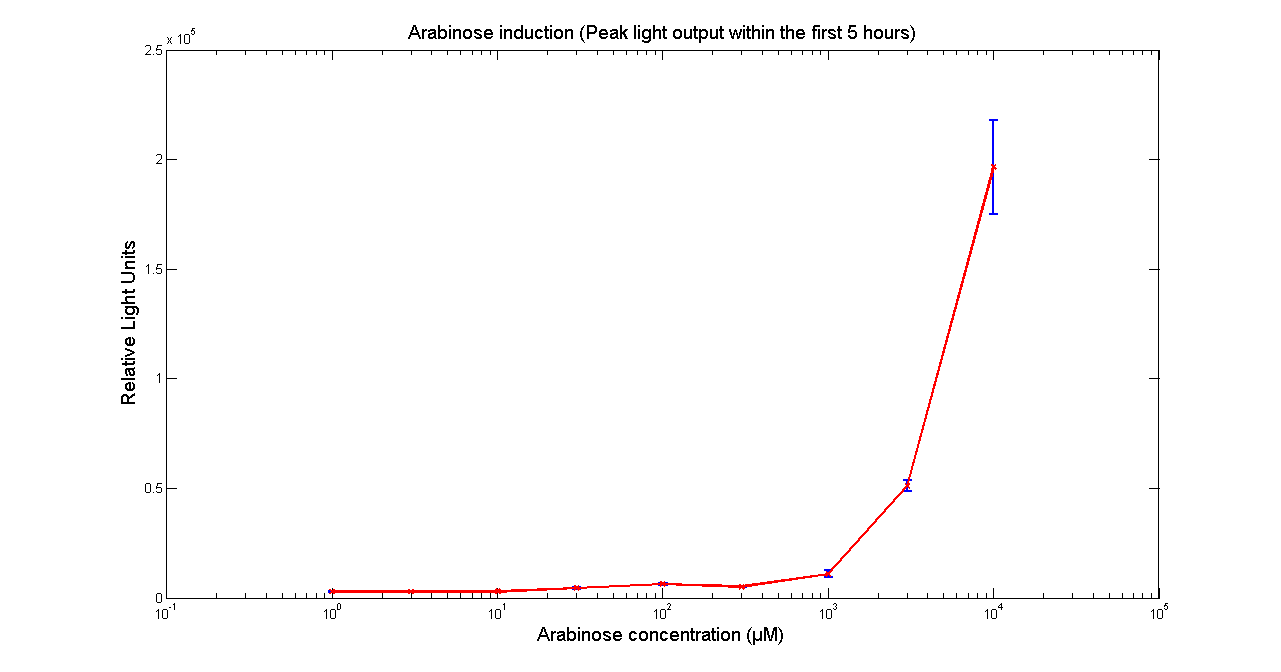
| - | ''' | + | [[Image:BBa_K325909AraK28.png|thumb|569px|center|'''Figure 1 - Peak light output from <partinfo>K325909</partinfo> cloned into H-NS mutant JM 230 H-NS -205::tn10. The data points and the error bar are the mean and standard deviation obtained by 3 tim repeats. ''']] |
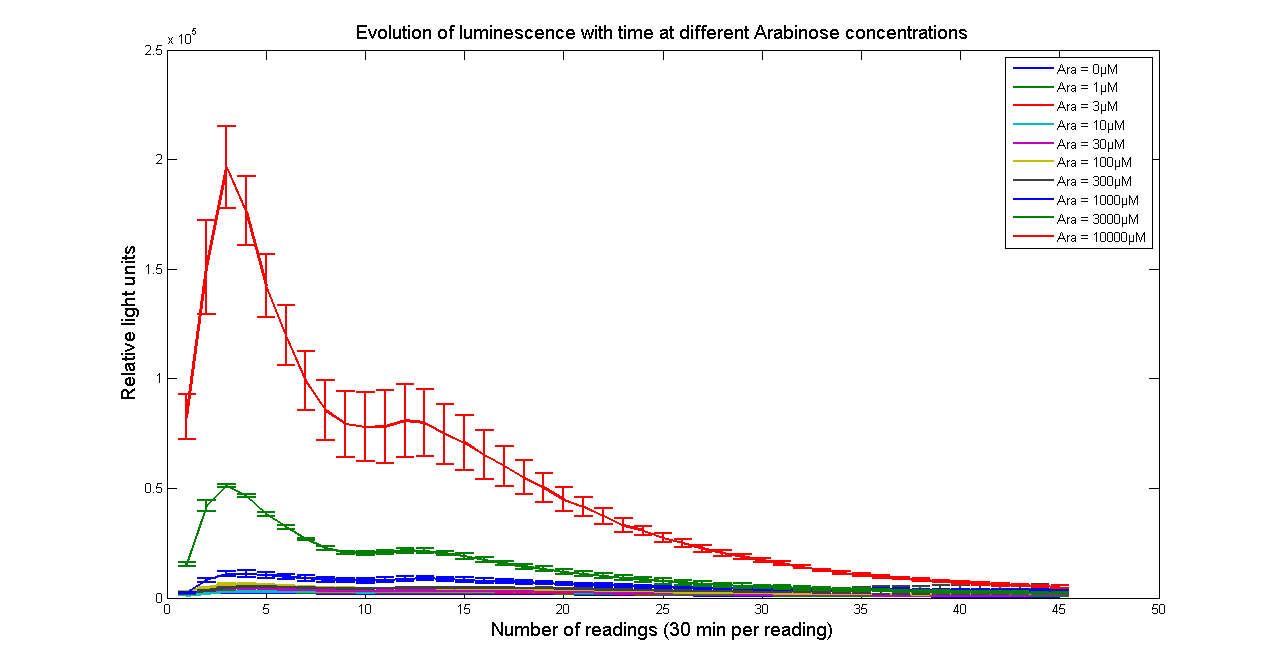
| - | + | [[Image:BBa_K325909timecourseK28.png|thumb|569px|center|'''Figure 2 - Evolution of light output from <partinfo>K325909</partinfo> cloned into H-NS mutant JM 230 H-NS -205::tn10 with time at different Arabinose concentrations. The data points and the error bar are the mean and standard deviation obtained by 3 time repeats. Measurements were taken every 30 minutes. ''']] | |
| - | + | [[Image:BBa_K325909AraR28.png|thumb|569px|center|'''Figure 3 - Figure 3 - Peak light output from <partinfo>K325909</partinfo> cloned into H-NS mutant [http://www.ecoliwiki.net/colipedia/index.php/BW25113 BW25113 DELTA H-NS::kan]. The data points and the error bar are the mean and standard deviation obtained by 3 tim repeats.''']] | |
| - | + | [[Image:BBa_K325909timecourseR28.png|thumb|569px|center|'''Figure 4 - Evolution of light output from <partinfo>K325909</partinfo> cloned into H-NS mutant [http://www.ecoliwiki.net/colipedia/index.php/BW25113 BW25113 DELTA H-NS::kan] with time at different Arabinose concentrations. The data points and the error bar are the mean and standard deviation obtained by 3 time repeats. Measurements were taken every 30 minutes.''']] | |
| - | The data points | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |- | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | '''Evolution of light output | + | |
| - | + | ||
| - | [http:// | + | |
| - | + | ||
| - | + | ||
<center> | <center> | ||
{|{{Table}} | {|{{Table}} | ||
| Line 102: | Line 53: | ||
!Date Uploaded | !Date Uploaded | ||
|- | |- | ||
| - | |[ | + | |[[Media:BBa_K325909Mutants.xls]] |
|Raw data from experiment | |Raw data from experiment | ||
|21/10/2010 | |21/10/2010 | ||
| Line 108: | Line 59: | ||
</center> | </center> | ||
| - | |||
| - | |||
=Compatibility= | =Compatibility= | ||
| - | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=cell ''Chassis:''] Device has been shown to work in ''Top 10 (Invitrogen)'' | + | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=cell ''Chassis:''] Device has been shown to work in ''Top 10 (Invitrogen)'', [http://www.ecoliwiki.net/colipedia/index.php/BW25113 BW25113 DELTA H-NS::kan] and H-NS mutant JM 230 H-NS -205::tn10.<br> |
[[Plasmid backbones|''Plasmids:'']] Device has been shown to work on ''<partinfo>pSB1C3</partinfo>'' <br> | [[Plasmid backbones|''Plasmids:'']] Device has been shown to work on ''<partinfo>pSB1C3</partinfo>'' <br> | ||
=References= | =References= | ||
| - | |||
| + | [http://www.jstor.org/stable/4449975 '''[1]:'''] J. Slock, (1995) Transformation Experiments Using Bioluminescence Genes of ''Vibrio fischeri'',''The American Biology Teacher'', '''57''', 225-227. | ||
| - | [http://www. | + | [http://www.annualreviews.org/doi/pdf/10.1146/annurev.mi.42.100188.001055 '''[2]:'''] E.A. Meighen (1988) Enzymes and genes from the ''lux'' operons of bioluminescent bacteria, ''Annual Reviews in Microbiology'' '''42''', 151-176. |
| + | [http://www.annualreviews.org/doi/pdf/10.1146/annurev.ge.28.120194.001001 '''[3]:'''] E.A. Meighen, (1994) Genetics of bacterial bioluminescence, ''Annual Reviews of Genetics'', '''28''', 117-139. | ||
| - | [http:// | + | [http://onlinelibrary.wiley.com/doi/10.1002/%28SICI%291099-1271%28199807/08%2913:4%3C185::AID-BIO486%3E3.0.CO;2-U/abstract '''[4]:'''] S. Ulitzur, (1998) H-NS controls the transcription of three promoters of ''Vibrio fischeri lux'' cloned in ''Escherichia coli'',''Journal of Bioluminescence and Chemiluminescence'', '''13'''(4), 185-188. |
| + | [http://www.nature.com/nature/journal/v444/n7117/full/nature05283.html '''[5]:'''] R.T. Dame ''et al.'', (2006) Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation,''Nature'', '''444''', 387-390. | ||
{{:Team:Cambridge/Templates/footer}} | {{:Team:Cambridge/Templates/footer}} | ||
Latest revision as of 23:24, 27 October 2010

This page describes characterisation for part BBa K325909, the lux operon from Vibrio fischeri.
Description

This page described the lux operon from Vibrio fischeri. To relieve LuxR control we placed Lux C, D, A, B, E under the pBad promoter.
Arabinose to light
This page describes the relationship between Arabinose concentration in the medium with light output. We used a FLUOstar OPTIMA microplate reader to quantify the light output. Protocol and plate reader settings used are given below.
Data
| Data | Notes | Date Uploaded |
|---|---|---|
| Media:BBa_K325909ArabinosetoLight.xls | Raw data from experiment | 21/10/2010 |
H-NS mutants
It has been shown that the expression of the Vibrio fischeri lux operon when cloned into E. coli was repressed. This repression was linked to the nucleoid protein H-NS. To investigate this effect we cloned the operon into mutant E.coli cells in which the expression of the H-NS protein had been modified. We used a FLUOstar OPTIMA microplate reader to quantify the light output. Protocols and plate reader settings used are given below.
Data
| Data | Notes | Date Uploaded |
|---|---|---|
| Media:BBa_K325909Mutants.xls | Raw data from experiment | 21/10/2010 |
Compatibility
Chassis: Device has been shown to work in Top 10 (Invitrogen), BW25113 DELTA H-NS::kan and H-NS mutant JM 230 H-NS -205::tn10.
Plasmids: Device has been shown to work on <partinfo>pSB1C3</partinfo>
References
[1]: J. Slock, (1995) Transformation Experiments Using Bioluminescence Genes of Vibrio fischeri,The American Biology Teacher, 57, 225-227.
[2]: E.A. Meighen (1988) Enzymes and genes from the lux operons of bioluminescent bacteria, Annual Reviews in Microbiology 42, 151-176.
[3]: E.A. Meighen, (1994) Genetics of bacterial bioluminescence, Annual Reviews of Genetics, 28, 117-139.
[4]: S. Ulitzur, (1998) H-NS controls the transcription of three promoters of Vibrio fischeri lux cloned in Escherichia coli,Journal of Bioluminescence and Chemiluminescence, 13(4), 185-188.
[5]: R.T. Dame et al., (2006) Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation,Nature, 444, 387-390.
 "
"