Team:Uppsala-SwedenWeek14
From 2010.igem.org
(Difference between revisions)
Syed.imtiyaz (Talk | contribs) |
Syed.imtiyaz (Talk | contribs) (→Construction Of K1, K2, K3) |
||
| Line 4: | Line 4: | ||
== Construction Of K1, K2, K3 == | == Construction Of K1, K2, K3 == | ||
| + | |||
| + | The samples inoculated previously were taken out and glecerol stocks were made. | ||
The inoclum obtained from the overnight culture was used for plasmid extraction. | The inoclum obtained from the overnight culture was used for plasmid extraction. | ||
| - | The extracted plasmids were cut at specific restriction sites present in the biobrick sequence and run on the gel. | + | The extracted plasmids were cut at specific restriction sites present in the biobrick sequence and run on the gel.` |
| - | The gel | + | The gel images of the bio-bricks with the band lengths were shown below in |
| - | + | Fig1, FIg2, Fig3 | |
| - | + | [[Image:20100906 K1 and K2 cPCR 20100902 (2).jpg|500px|center|border|thumb|K1-K2 Figure 1]] | |
| - | + | ||
| - | + | ||
| - | + | [[Image:20100906 K1 and K2 cPCR 20100902 thumb.jpg|500px|center|border|thumb|K1-K2 Figure 2]] | |
| - | + | [[Image:20100906 K3 cPCR 20100902 (2) thumb.jpg|500px|center|border|thumb|K3 Figure 3]] | |
| - | The | + | |
| + | The gel was checked for correct fragment lengths to perform a second round f validation | ||
| + | The extracted plasmids were digested with the correct set of enzymes as defined in the flow chart. | ||
| + | |||
| + | The concentration of plasmid were measured for all the samples. | ||
Latest revision as of 21:12, 26 October 2010



Week-14
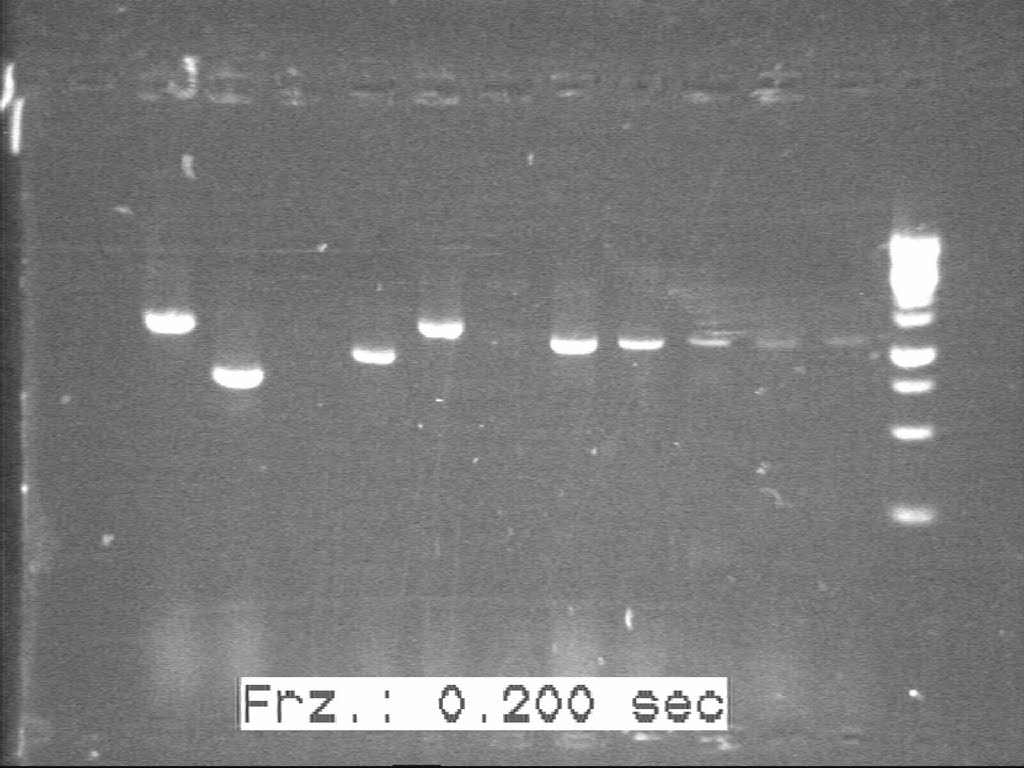
Construction Of K1, K2, K3
The samples inoculated previously were taken out and glecerol stocks were made.
The inoclum obtained from the overnight culture was used for plasmid extraction. The extracted plasmids were cut at specific restriction sites present in the biobrick sequence and run on the gel.`
The gel images of the bio-bricks with the band lengths were shown below in Fig1, FIg2, Fig3
The gel was checked for correct fragment lengths to perform a second round f validation
The extracted plasmids were digested with the correct set of enzymes as defined in the flow chart.
The concentration of plasmid were measured for all the samples.
 "
"