Team:Washington/Tools Created/New Vectors
From 2010.igem.org
(→f1 origin) |
|||
| (85 intermediate revisions not shown) | |||
| Line 25: | Line 25: | ||
</html> | </html> | ||
<!---------------------------------------PAGE CONTENT GOES BELOW THIS----------------------------------------> | <!---------------------------------------PAGE CONTENT GOES BELOW THIS----------------------------------------> | ||
| - | = | + | == Protein Expression Vectors == |
| - | + | ==[[Image:Washington_2010_vector_logo2.jpg|center|250px|]]== | |
| - | + | ||
=='''Vector Design'''== | =='''Vector Design'''== | ||
| - | [[Image: | + | [[Image:Washington_2010_vector7.jpg|center|750px|]] |
| - | + | <br /> | |
| - | |||
| - | + | [[Image:uw_vectors_button.jpg|150px|left]] | |
| - | The expression | + | <br /> |
| + | <br /> | ||
| + | The protein expression cassette is designed to go into any biobrick plasmid backbone. It was placed in 4 different plasmid backbone from the registry [http://partsregistry.org/Part:pSB1C3 pSB1C3], [http://partsregistry.org/Part:pSB1A3 pSB1A3], [http://partsregistry.org/Part:pSB3K3, pSB3K3],and [http://partsregistry.org/Part:pSB4A5 pSB4A5] by our lab. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| - | + | [[Image:uw_promoters_button3.jpg|140px|left]] | |
| + | <br /> | ||
| + | <br /> | ||
| + | The expression vector promoters are available in constitutive and inducible variety. The [http://partsregistry.org/Part:BBa_J23100 Constitutive promoters] are BBa J23100 and J23114. Inducible vectors include [http://partsregistry.org/Part:BBa_R0011 R0011] and [http://partsregistry.org/Part:BBa_K314112 T7], both of which are repressed by the Lac I protein. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | [[Image:uw_Lac_button.jpg|140px|left]] | ||
| + | <br /> | ||
| + | Lac I gene inserted to ensure regulation of the Lactose inducible promoters | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | [[Image:uw_AB_button.jpg|140px|left]] | ||
| + | <br /> | ||
| + | Antibiotics resistance based on biobrick plasmid backbone used. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | [[Image:uw_RBS_button.jpg|140px|left]] | ||
| + | <br /> | ||
The Elowitz standard RBS [http://partsregistry.org/Part:BBa_B0034 B0034] is used on all vectors. | The Elowitz standard RBS [http://partsregistry.org/Part:BBa_B0034 B0034] is used on all vectors. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| - | + | [[Image:uw_F1_button.jpg|140px|left]] | |
| - | + | Origin of replication for the M13 series of phages. When included in plasmids the cells can be infected by M13 phage which will then replicate and package the plasmid as single stranded DNA. This DNA can then be isolated as described in our [[Team:Washington/Protocols/KunkelCapD|Kunkel Mutagenesis protocol]]. | |
| - | + | <br /> | |
| + | <br /> | ||
| + | [[Image:uw_copy_button.jpg|140px|left]] | ||
| + | <br /> | ||
| + | Plasmid copy number based on biobrick plasmid backbone used. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | [[Image:uw_cut_button.jpg|100px|left]] | ||
| + | <br /> | ||
| + | <br /> | ||
| + | Restriction cut sites based on biobrick standards. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | =='''Building the Vectors'''== | ||
| + | The expression cassettes were built using the [http://openwetware.org/wiki/BioBricks_construction_tutorial biobricks construction tutorial]. | ||
=='''Testing the Vectors'''== | =='''Testing the Vectors'''== | ||
| - | ===''' | + | ==='''Protein Expression'''=== |
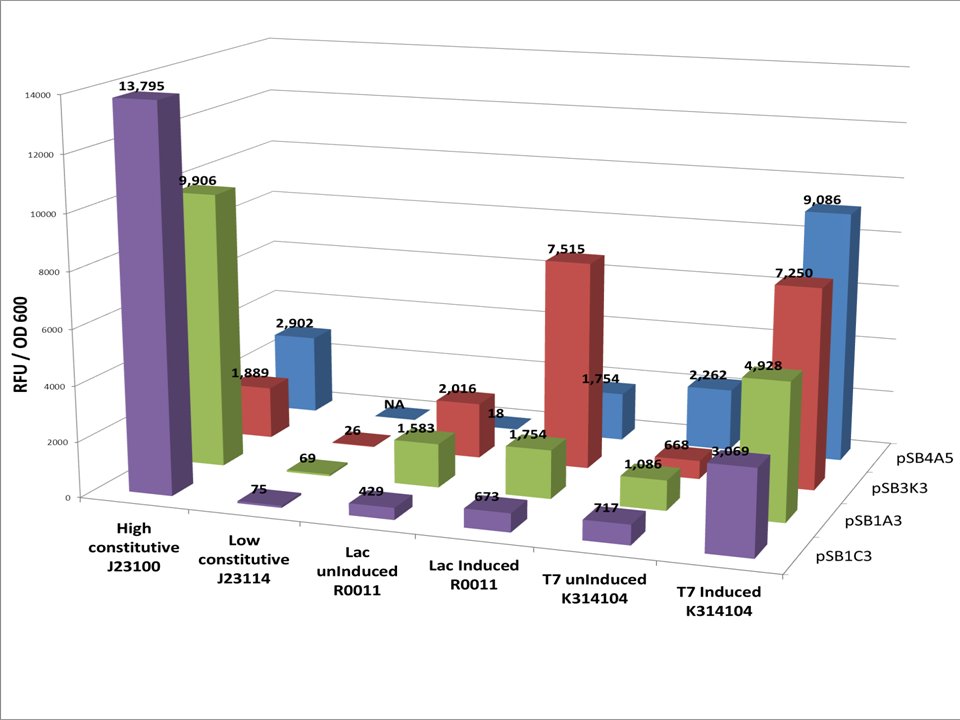
| - | The vector | + | The vector cassettes was placed in 4 different plasmid backbones from the registry [http://partsregistry.org/Part:pSB1C3 pSB1C3], [http://partsregistry.org/Part:pSB1A3 pSB1A3], [http://partsregistry.org/Part:pSB3K3, pSB3K3], [http://partsregistry.org/Part:pSB4A5 pSB4A5] and [http://partsregistry.org/Part:BBa_E0040 GFP] was placed in the protein expression area of the vector. The expression cassette was grown according to our [[Team:Washington/Protocols/VectorAssay|vector assay]] protocol. Data was corrected for background florescence. |
| - | [[Image: | + | [[Image:Washington2010_vector_assay_data.png|thumb|left|650px|'''Figure 1. GFP fluorescence was measured (ex. 485, em. 525) and normalized to cell density, read as the optical density at 600nm.''']] |
| + | <br style="clear: both" /> | ||
==='''f1 origin'''=== | ==='''f1 origin'''=== | ||
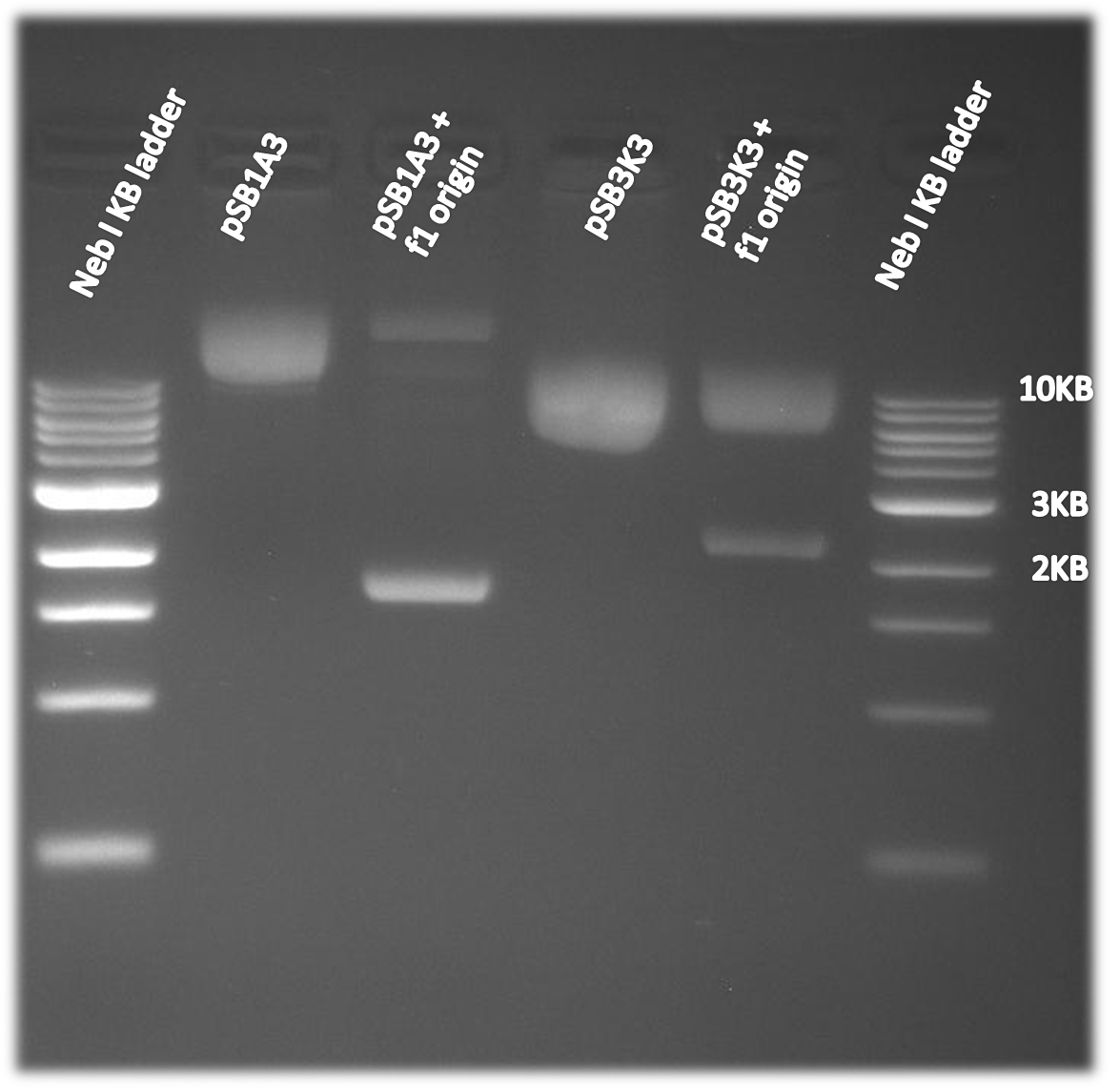
| - | The f1 origin was tested by | + | The f1 origin was tested by comparing single strand DNA harvest using part of the [https://2010.igem.org/Team:Washington/Protocols/KunkelCapD Kunkel Mutagenesis]. CJ236 cells were infected with M13K07 helper phage. The CJ236 cells varied in the present or absence of the f1 origin on the pSB1A3 or pSB3K3 plasmid. The single stranded DNA was harvested and then run on a 1% agarose gel. As is clearly visible on the gel below single strand DNA of the plasmid (~2-3kb, depending on the plasmid) is only made when the f1 origin is present. Note that single stranded DNA runs at a slightly smaller than expected size when comparing to a double stranded DNA ladder. |
| - | [[Image: | + | |
| + | [[Image:Washington_f1_origin_gel.png|thumb|left|400px|'''Figure 2. Single stranded DNA harvested from phage and run on a gel. The higher bands are phage genomic DNA while the lower bands at roughly 2-3kb correspond to the expected plasmid size.''']] | ||
| + | <br style="clear: both" /> | ||
| + | |||
| + | |||
| + | |||
<!---------------------------------------PAGE CONTENT GOES ABOVE THIS----------------------------------------> | <!---------------------------------------PAGE CONTENT GOES ABOVE THIS----------------------------------------> | ||
<div style="text-align:center"> | <div style="text-align:center"> | ||
| - | '''← [[Team:Washington | + | '''← [[Team:Washington/Tools Created|Overview of Tools We Created]]''' |
| | ||
| | ||
| | ||
| - | '''[[Team:Washington/ | + | '''[[Team:Washington/Tools Created/New Software|New Software]] →''' |
</div> | </div> | ||
{{Template:Team:Washington/Templates/Footer}} | {{Template:Team:Washington/Templates/Footer}} | ||
Latest revision as of 00:12, 28 October 2010
Protein Expression Vectors
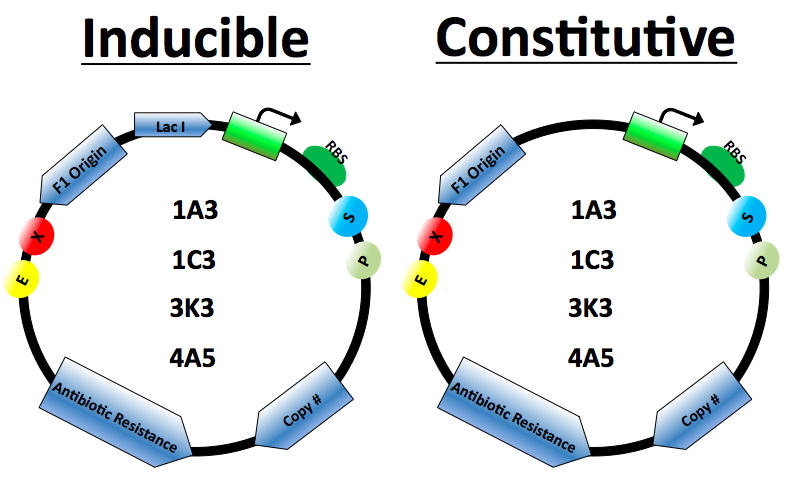
Vector Design
The protein expression cassette is designed to go into any biobrick plasmid backbone. It was placed in 4 different plasmid backbone from the registry [http://partsregistry.org/Part:pSB1C3 pSB1C3], [http://partsregistry.org/Part:pSB1A3 pSB1A3], [http://partsregistry.org/Part:pSB3K3, pSB3K3],and [http://partsregistry.org/Part:pSB4A5 pSB4A5] by our lab.
The expression vector promoters are available in constitutive and inducible variety. The [http://partsregistry.org/Part:BBa_J23100 Constitutive promoters] are BBa J23100 and J23114. Inducible vectors include [http://partsregistry.org/Part:BBa_R0011 R0011] and [http://partsregistry.org/Part:BBa_K314112 T7], both of which are repressed by the Lac I protein.
Lac I gene inserted to ensure regulation of the Lactose inducible promoters
Antibiotics resistance based on biobrick plasmid backbone used.
The Elowitz standard RBS [http://partsregistry.org/Part:BBa_B0034 B0034] is used on all vectors.
Origin of replication for the M13 series of phages. When included in plasmids the cells can be infected by M13 phage which will then replicate and package the plasmid as single stranded DNA. This DNA can then be isolated as described in our Kunkel Mutagenesis protocol.
Plasmid copy number based on biobrick plasmid backbone used.
Restriction cut sites based on biobrick standards.
Building the Vectors
The expression cassettes were built using the [http://openwetware.org/wiki/BioBricks_construction_tutorial biobricks construction tutorial].
Testing the Vectors
Protein Expression
The vector cassettes was placed in 4 different plasmid backbones from the registry [http://partsregistry.org/Part:pSB1C3 pSB1C3], [http://partsregistry.org/Part:pSB1A3 pSB1A3], [http://partsregistry.org/Part:pSB3K3, pSB3K3], [http://partsregistry.org/Part:pSB4A5 pSB4A5] and [http://partsregistry.org/Part:BBa_E0040 GFP] was placed in the protein expression area of the vector. The expression cassette was grown according to our vector assay protocol. Data was corrected for background florescence.
f1 origin
The f1 origin was tested by comparing single strand DNA harvest using part of the Kunkel Mutagenesis. CJ236 cells were infected with M13K07 helper phage. The CJ236 cells varied in the present or absence of the f1 origin on the pSB1A3 or pSB3K3 plasmid. The single stranded DNA was harvested and then run on a 1% agarose gel. As is clearly visible on the gel below single strand DNA of the plasmid (~2-3kb, depending on the plasmid) is only made when the f1 origin is present. Note that single stranded DNA runs at a slightly smaller than expected size when comparing to a double stranded DNA ladder.
 "
"