Team:Debrecen-Hungary/project
From 2010.igem.org
(→Contibution to project by team members) |
|||
| (96 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Template:Debrecen- | + | {{Template:Debrecen-Hungary_secret}} |
| Line 5: | Line 5: | ||
<html> | <html> | ||
| - | <table width="910"> | + | <table width="910" bgcolor="#EDE8E2" border="0" cellspacing="0" cellpadding="0"> |
<tr> | <tr> | ||
| - | <td align="left"> | + | <td align="left" bgcolor="#EDE8E2" border="0" cellspacing="0" cellpadding="0"> |
| - | <img src=" | + | <img src="https://static.igem.org/mediawiki/2010/e/ee/Projectbck.jpg"> |
</td> | </td> | ||
| - | <td align="right"> | + | <td align="right" bgcolor="#EDE8E2" border="0" cellspacing="0" cellpadding="0"> |
| - | <a href="https://2010.igem.org/Team:Debrecen-Hungary"><img src=" | + | <a href="https://2010.igem.org/Team:Debrecen-Hungary"><img src="https://static.igem.org/mediawiki/2010/9/96/Backbuttonbck.jpg" align="right"></a> |
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
<br> | <br> | ||
| + | <a name="abs"></a><br></html> | ||
| + | ='''Abstract'''= | ||
| - | + | Eukaryotic synthetic biology has huge potential, yet it is still in need of more diverse molecular tools for defined gene regulation. | |
| - | + | Nuclear receptors are a conserved family of proteins responsible for sensing lipids; they may be viewed as lipid activated transcription factors. | |
| - | + | '''We have successfully developed a kit with a variety of lipid responsive domains''' (from H.sapiens, D.melanogaster and C.elegans) '''for the rational construction of synthetic transcription factors.''' The domains respond only to predefined lipids and selectively activate predetermined gene expression. | |
| + | To characterize theses domains, we used standardized protocols for comparable measurements. In vivo gene expression was measured as a function of ligand concentration using luciferase activity. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | '''The potential for these tools is immense; e.g. from the ultra sensitive detection of lipid contaminants in the environment to the opportunity of titration specific gene expression canges in patients undergoing gene therapy. Regarding applications in food quality control assessment, these system allows the measurement of Vitamin A, Vitamin A derivatives through RAR (retinoic acid receptor), vitamin D and its derivatives through VDR (vitamin D receptor), but also the detection of steroids (antiinflamatory steroids, anabolic steroids, estrogens) through GR (glucocorticoid receptor), ER (estrogen receptors), AR (testosteron receptor). By simply copying the xenobiotic receptors of humans (e.g. PXR, CAR) we can assess the presence of antibiotics in food or wheter existing or new food additives will activate our endogenous xenobiotic system. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | '''In summary the Nuclear Receptor Toolkit developped by us allows the (1) ultra sensitive detection of xenobiotic activators in the environment (e.g. oil derived traces in water and soil), (2) the assessment of lipid soluble vitamins, steroids, xenobiotics in food and (3) the potential of inducing specific genetic programs in transfected cells or modell organisms with a possibility to destroy these modified cells by an independent tetracyclin induced apoptosis effector system.''' ''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <html><br><a name="intro"></a><br></html> |
| - | + | ||
| - | + | ||
| - | + | ='''Introduction'''= | |
| - | + | ==''Synthetic Biology''== | |
| - | + | ||
| - | + | ||
| - | + | '''Synthetic biology''' is a relatively new area of biological research; similar to many other new scientific fields it has many definitions. The best way to express the meaning of synthetic biology is to understand the desired end-result: ''engineering of complex biological systems''. These systems are best thought of as analogues of everyday machinery: cogwheels, levers, timers, button and buzzers (in this case a clock), only in the case of biological systems (molecular ones) cells, DNA, proteins, lipids, sugars and RNA are the “parts” of the system. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Similar to mechanical engineering (or every other engineering branch) there is a need for standards (consensus way of doing things), abstraction (simple and unified way of thinking about the parts of a system), and modularity (how these parts interact to become the device, or several devices into a system). Thus a good definition for synthetic biology could be engineering of molecular (for the time being) biological systems '''according to preset standard parts'''. | |
| - | + | The international genetically engineered machines and associated parts registry are, to date, one of the largest registries for standard parts in use for synthetic biology. Free information can be found in the registry regarding parts, devices and modules all inputted by various teams worldwide. | |
| - | + | ||
| - | + | ||
| - | + | [https://2010.igem.org/Team:Debrecen-Hungary/minimals#Drosophila_Melanogaster Eukaryotic synthetic biology] is still in its infancy. The large kingdom of metazoans includes all multicellular eukaryotes such as mammalians, arthropods and nematodes. No standard chassis (framework) exists for the animal kingdom which makes them far less popular then the famous E.coli. The protein modules derived from metazoans (like Drosophila or C. elegans) are functional in yeasts also. Very few iGEM teams (or even labs outside iGEM) have chosen to toggle the animal chassis (two notable examples are team Heidelberg 2009 and team Slovenia 2006). The amount of available compatible parts is limited, which severely restricts the options of creating complex biological devices. Nearly no imagination is required for designing tools, since their analogues already exist in the bacterial chassis. The possible use of such systems is unlimited. Field’s such as of environment, medicine, energy and research all gain to profit from the development of animal synthetic biology. | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | Systems requiring gene expression input in eukaryotic synthetic biology systems require a way to standardize gene expression, a complicated task. The way from gene to protein contains many steps of possible error: transcription factor binding, promoter strength, recruitment of auxiliary proteins, nuclear RNA synthesis and many more steps finally leading to translation, folding, cleaving and delivery (but hey, you have to start somewhere). | ||
| - | + | '''Our team was interested at designing eukaryotic synthetic biology tools related to PoPs'''. PoPs (polymerase per second), the flying Dutchman of synthetic biology, is a number which represents the rate (base pair per second) at which RNA polymerase crosses past a given DNA position. Currently, no in vivo technique for measuring PoPS directly exists; it can be estimated indirectly by measuring other parameters (eg protein expression or enzyme activity). Nevertheless it is still a useful abstraction for thinking about transcription-based logic devices and it allows the engineer to define devices. Our aim, was not only to infer PoPs but to devise a way to titrate it remotely. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | ==''New Tools for Synthetic Biology: Nuclear Receptors''== |
| - | <a name=" | + | |
| - | < | + | [https://2010.igem.org/Team:Debrecen-Hungary/minimals#Scientific_Background Nuclear receptors] (click to find out about nuclear receptors), best viewed as transcription factors which can be activated by extracellular cues, are unique in their ability to allow direct remote PoPs titration (both activation and repression). These receptor classes bears high homology to each other throughout the animal kingdom and are modular into distinct domains. All of these features attracted our attention to find a way to incorporate these tools in the parts registry, and characterize them in standardized methods. |
| - | <br> | + | |
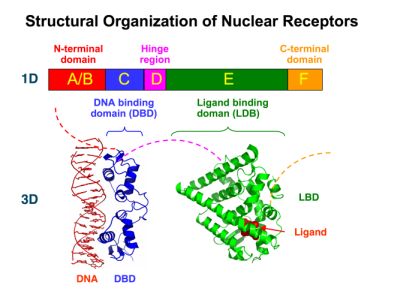
| + | Many nuclear receptors, are among the primary targets of drug discovery because of their diverse biological actions. Nuclear receptors bear high homology to each other and are modular into distinct domains: N-terminal regulatory domain, DNA-binding domain (DBD), a Hinge region, Ligand binding domain (LBD) and a C-terminal domain. [https://2010.igem.org/Team:Debrecen-Hungary/minimals#Scientific_Background The LBD] (click to find out more about LBD's) is the segment of the receptor which changes its conformation upon lipophilic ligand binding; this also causes the exposure of a powerful transcriptional activation domain (which attracts the transcriptional machinery). This was our segment of interest since it links ligand binding with transcriptional activity. | ||
| + | |||
| + | [[Image:Nuclear_Receptor_Structure_resized.jpg]] | ||
| + | |||
| + | '''Our team has generated a library of ligand binding domains''' for the rational construction of synthetic titrateable transcription factors. In our model hybrid receptor, this LBD was '''fused to a Gal4 DNA binding domain'''. The sequence to which Gal4 binds is CGG-N11-CCG, where N can be any base. Although Gal4 is a yeast protein not normally present in other organisms it has been shown to work as a transcription factor in a variety of organisms such as Drosophila, and human cells, highlighting that the same mechanisms for gene expression have been conserved over the course of evolution. | ||
| + | |||
| + | The examination of these chimeric receptors was through a version of the [https://2010.igem.org/Team:Debrecen-Hungary/minimals#Cotransfection_Assay_Two_Hybrid_Assay two hybrid assay] (click to find out more about the two hybrid assay). For a more detailed description of our experimental system, please visit our [https://2010.igem.org/Team:Debrecen-Hungary/minimals#Cotransfection_Assay_Two_Hybrid_Assay "Minimals"] page. | ||
| + | |||
| + | <html><br><a name="results"></a><br></html> | ||
| + | |||
| + | ='''Results'''= | ||
| + | |||
| + | First we had to '''generate expression vectors''' that can be used in mammalian expression systems. The cloning work of Team Debrecen-Hungary resulted [https://2010.igem.org/Team:Debrecen-Hungary/parts#Ligand_binding_domains_-_Basic_Parts 18 basic parts] in pSB1C3 and [https://2010.igem.org/Team:Debrecen-Hungary/parts#Chimeric_Nuclear_Receptors_-_Composite_Parts 13 composite parts] in pSB1C3 or pSB1A3. All of them have been tested by restriction enzyme analysis and by sequencing. Furthermore, some basic and fused parts have been ligated into pCDNA3.1 containing a specific adaptor by which RFC-25 compatible parts can be inserted into this expression vector. pCDNA3.1 results constitutive expression of the inserted part in the cells so the investigated part can be tested at functional level. The other [https://2010.igem.org/Team:Debrecen-Hungary/parts#Eukaryotic_expression_vectors_-_Composite_Parts expression vector] is a fully SB (Synthetic Biology) compatible mammalian expression vector generated from pSB1A3. We inserted into the pSB1A3 the TRE (tetracycline response element) a strong CMV (cytomegalo virus) promoter and a PolyA element that is a terminator of transcription in mammalian cells. These four elements are available as individual parts in the parts registry and can be used in combination with other parts too. | ||
| + | |||
| + | =='''Cloning'''== | ||
| + | |||
| + | ===Basic Parts=== | ||
| + | |||
| + | [https://2010.igem.org/Team:Debrecen-Hungary/parts#Ligand_binding_domains_-_Basic_Parts '''18 basic parts'''] have been inserted into pSB1C3 which contain 3 DBDs, 1 hinge, 11 LBDs, 2 regulators and 1 apoptosis protein. A Gal4 DBD which has the RFC25 N terminal linkers was designed and submitted to be used (BBa_K364319). | ||
| + | |||
| + | ===Composite Parts=== | ||
| + | |||
| + | [https://2010.igem.org/Team:Debrecen-Hungary/parts#Chimeric_Nuclear_Receptors_-_Composite_Parts '''13 composite parts'''] have been created, ten of them consist of two basic parts, one of them has been built up from three basic parts, and an expression vector from standard biobrick parts has been created by using four basic parts. The backbone has been opened with AgeI and PstI restriction enzymes in order to ligate the part cut out with NgoMIV and PstI. The previous restriction digests have been repeated so each part could be inserted after the previous one. | ||
| + | |||
| + | ====Gal4-human LBD fusions==== | ||
| + | |||
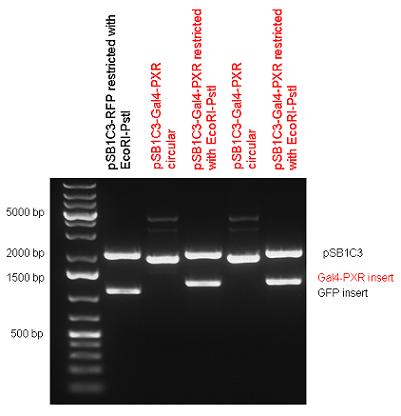
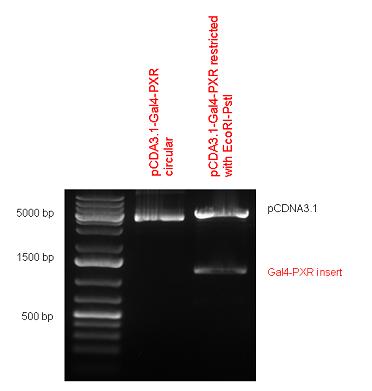
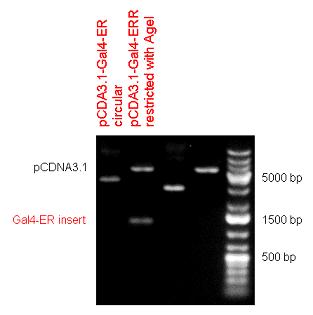
| + | Two human nuclear hormone receptor LBDs (Pregnane X Receptor LBD, Estrogen Receptor LBD) have been inserted into pSB1C3 after Gal4 was ligated into the backbone. The Gal4 (438 bp) is a modular protein consisting of a DNA-binding domain and an activation domain which can be used in [https://2010.igem.org/Team:Debrecen-Hungary/minimals#Cotranfection_Assay,_Two_Hybrid_Assay Two-Hybrid system] in order to identify interactions between two proteins or a protein with DNA. The length of PXR LBD is 732 bp while the ER LBD is 828 bp long, so the fusion leads to 1170 bp and 1266 bp long composite inserts. | ||
| + | |||
| + | {| | ||
| + | | [[Image:Fig._1._Team_Debrecen_pSB1C3-Gal4-PXR.JPG|500px|thumb|upright|alt=|pSB1C3-Gal4-PXR]] | ||
| + | | [[Image:Fig.2.Team_Debrecen_pSB1C3-Gal4-ER.JPG|500px|thumb|upright|alt=|pSB1C3-Gal4-ER]] | ||
| + | |} | ||
| + | |||
| + | Both of them have been transferred into pCDNA3.1 to express them in certain cell lines. | ||
| + | |||
| + | {| | ||
| + | | [[Image:Fig_3.Team_Debrecen_pCDNA-Gal4-PXR.JPG|500px|thumb|upright|alt=|pCDNA-Gal4-PXR]] | ||
| + | | [[Image:Fig._4._Team_Debrecen_pCDNA-Gal4ER.JPG|500px|thumb|upright|alt=|pCDNA-Gal4ER]] | ||
| + | |} | ||
| + | |||
| + | ====Gal4-Ecdysone receptor fusion==== | ||
| + | |||
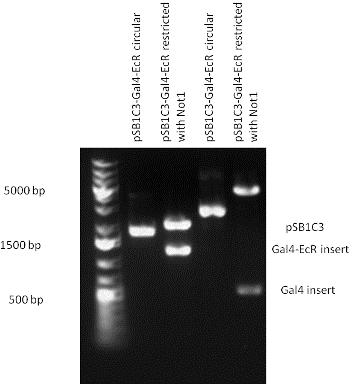
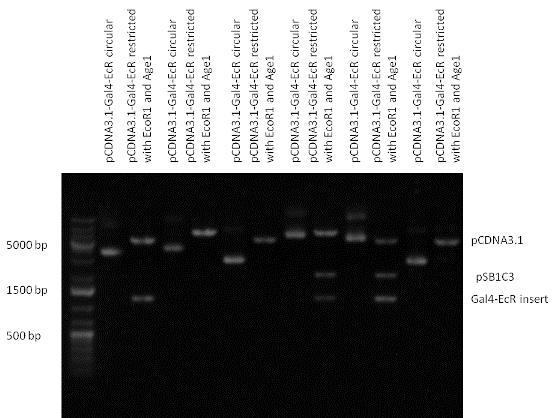
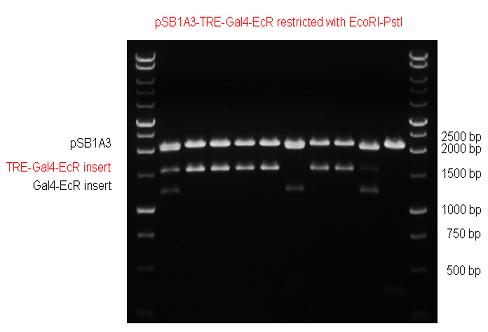
| + | The LBD of Ecdysone Receptor (639 bp) which can be found in arthropodes has been inserted after the Gal4 both in the pSB1C3 and pCDNA3.1 resulting an 1077 bp long insert in the vectors. | ||
| + | |||
| + | {| | ||
| + | | [[Image:Fig._5._Team_Debrecen_PSB1C3-Gal4-EcR.JPG|500px|thumb|upright|alt=|pSB1C3-Gal4-EcR]] | ||
| + | | [[Image:Fig._6._Team_Debrecen_PCDNA-Gal4-EcR.JPG|500px|thumb|upright|alt=|pCDNA-Gal4-EcR]] | ||
| + | |} | ||
| + | |||
| + | ====Gal4-C. elegans LBD fusions==== | ||
| + | |||
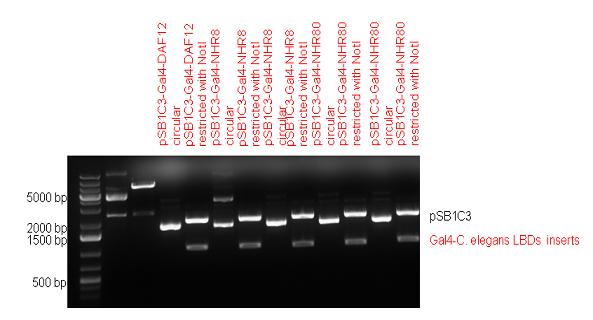
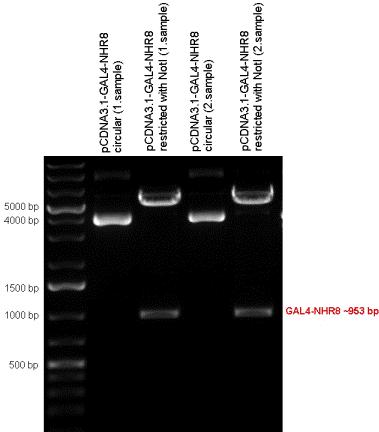
| + | Five nuclear hormone receptor LBDs of C. elegans (NHR8, NHR23, NHR31, NHR80, DAF12) have been inserted after Gal4 in pSB1C3, resulting inserts with the lenght between 944 bp and 977 bp. All of the Gal4-C.elegans LBD constructs (e.g. Gal4-NHR8) except Gal4-NHR80 composite part have been fused into pCDNA3.1 expression vector to test them as a lipid sensor. | ||
| + | {| | ||
| + | | [[Image:Fig._7._Team_Debrecen_PSB1C3-Gal4-NHR.JPG|500px|thumb|upright|alt=|pSB1C3-Gal4-NHR]] | ||
| + | | [[Image:Fig._8._Team_Debrecen_PCDNA-Gal4-NHR8.JPG|500px|thumb|upright|alt=|pCDNA-Gal4-NHR8]] | ||
| + | |} | ||
| + | |||
| + | ====Estrogen Receptor from basic parts==== | ||
| + | |||
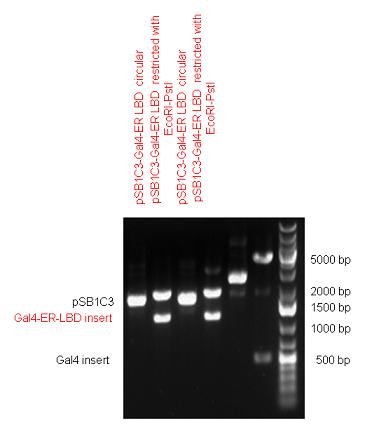
| + | Estrogen receptor DBD (318 bp) and LBD (828 bp) have been inserted together into pSB1C3. The two main domains of ER are linked together by the hinge region of Vitamin D Receptor (357 bp) resulting a 1503 bp long insert in the backbone. | ||
| + | {| | ||
| + | | [[Image:Fig.9.Team_Debrecen_pSB1C3-ER_DBD-VDRh-ER_LBD.JPG|500px|thumb|upright|alt=|pSB1C3-ER_DBD-VDRh-ER_LBD]] | ||
| + | |} | ||
| + | |||
| + | ===Creating expression vector from pSB1A3=== | ||
| + | |||
| + | Three basic parts from pSB1C3 have been ligated into pSB1A3 consecutively by using chloramphenicol and ampicillin assembly in order to create an [https://2010.igem.org/Team:Debrecen-Hungary/parts#Eukaryotic_expression_vectors_-_Composite_Parts expression vector] from the ampicillin resistance carrying backbone. | ||
| + | {| | ||
| + | | [[Image:Fig._10._Team_Debrecen_pSB1A3-TRE-Gal4-PXR.JPG|450px|thumb|upright|alt=|pSB1A3-TRE-Gal4-PXR]] | ||
| + | | [[Image:Fig._11._Team_Debrecen_pSB1A3-TRE-Gal4-EcR.JPG|450px|thumb|upright|alt=|pSB1A3-TRE-Gal4-EcR]] | ||
| + | |} | ||
| + | |||
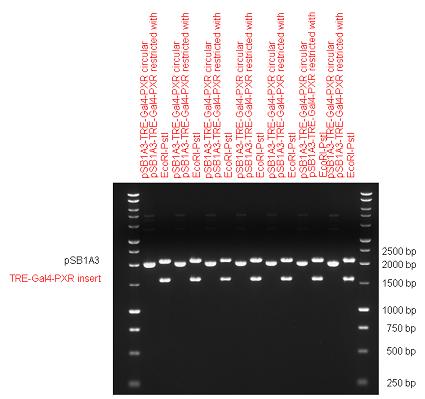
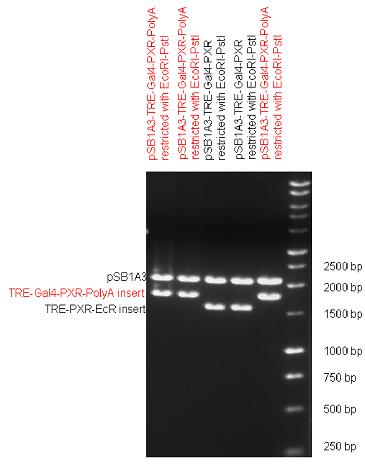
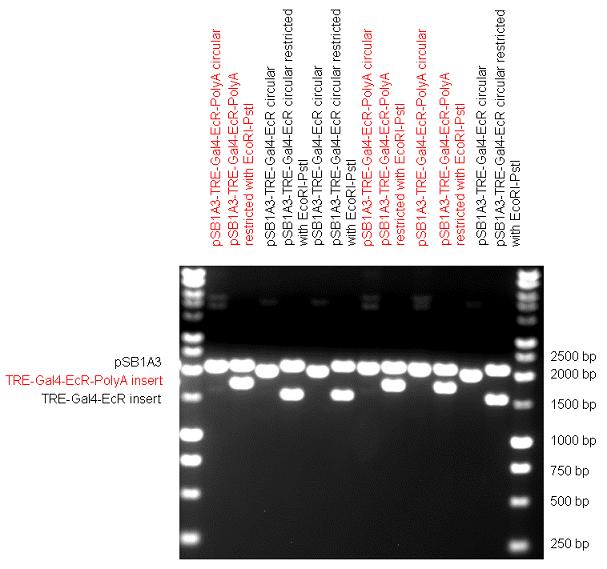
| + | The first step was to insert 321 bp long TRE-CMV (Tetracyclin Response Element- Minimal Cytomegalovirus promoter) into pSB1A3 which has the length of 2157 bp. Then Gal4-PXR LBD (1170 bp) and Gal4-Ecr LBD (1077 bp) fusions were inserted after the TRE- CMV, before PolyA tail (191 bp) was ligated in the end. | ||
| + | {| | ||
| + | | [[Image:Fig._12._Team_Debrecen_pSB1A3-TRE-Gal4-PXR-PolyA.JPG|450px|thumb|upright|alt=|pSB1A3-TRE-Gal4-PXR-PolyA]] | ||
| + | | [[Image:Fig_13._Team_Debrecen_pSB1A3-TRE-Gal4-EcR-PolyA.JPG|450px|thumb|upright|alt=|pSB1A3-TRE-Gal4-EcR-PolyA]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | As the negative control of the expression vector built up from standard biobrick parts TRE- CMV and PolyA tail were ligated together into pSB1C3 and pSB1A3. | ||
| + | {| | ||
| + | | [[Image:Fig_14._Team_Debrecen_pSB1A3-TRE-PolyA.JPG|450px|thumb|upright|alt=|pSB1A3-TRE-PolyA]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | ===Western Blots of Gal-fused proteins=== | ||
| + | |||
| + | The functionality of the constructs was assesed by Gal4 '''Western Blot'''. | ||
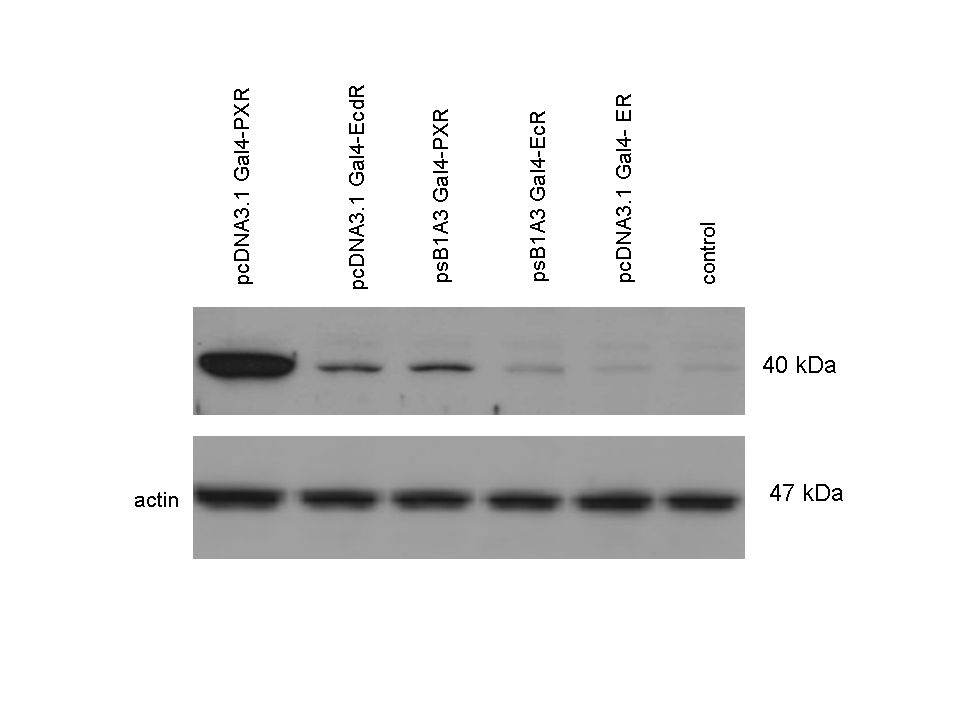
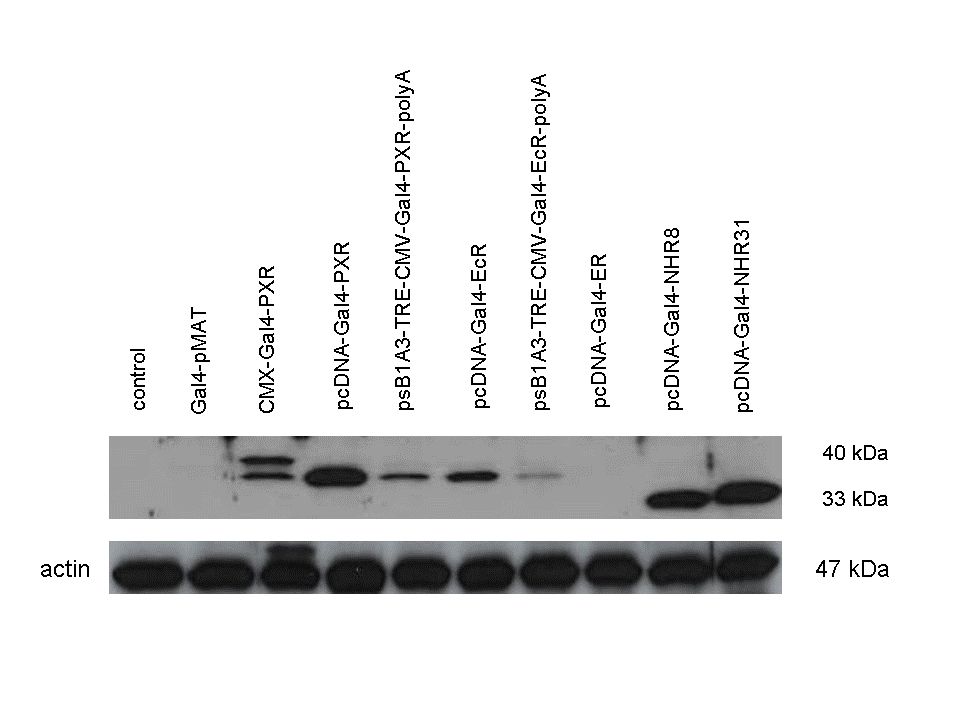
| + | 293T cells were transfected. Western-blot was carried out using '''rabbit anti-Gal4 polyclonal''' primary antibody. High protein expression have been noticed when Gal4-PXR LBD composite construct was cloned into pCDNA3.1 as well as into the expression vector created from pSB1A3 (using standard BioBrick parts). Lower expression was detected in the case of Gal4-EcR LBD in both expression vectors. The Gal4-PXR and EcR-LBD fused proteins appeared at about '''40 kDa''' while the Gal4-C.elegans-LBD (NHR8, NHR31) chimeric proteins which have been intensely expressed, can be seen between '''30-35 kDa''' which corresponds to thier predicted molecular weight. | ||
| + | |||
| + | {| | ||
| + | | [[Image:IGEM_WB-1.jpg|450px|thumb|upright|alt=|]] | ||
| + | | [[Image:IGEM_WB-2.jpg|450px|thumb|upright|alt=|]] | ||
| + | |} | ||
| + | |||
| + | ==Transients cotransfection assay /two-hybrid system/== | ||
| + | |||
| + | |||
| + | We '''moved the composite parts generated in pSB1C3 into the expression vectors''', then transfered these fused parts into pCDNA and pSB1A3 vectors for further works. pCDNA is designed for high-level expression, purification and detection of recombinant protein in mammalian hosts. In addition we developed a new expression vector pSB1A3 with a CMV-promoter and poly-A tail. Our experiments proceeded on two different lines. | ||
| + | |||
| + | The first one is the presentation of the cloned constructs bearing the '''ability to get activated''' by ligand binding. These were the activation experiments. | ||
| + | |||
| + | Furthermore we showed that our constructs had the '''ability to interact with the previously published heterodimeric partner RXR'''. To confirm this we used the mammalian two-hybrid system. Binding to RXR confirms the proper folding and ultimately the generation of a well designed LBD, and a functional mammalian expression vector. The method used to design our parts is described as a proposed standard BBF RFC 64 [https://static.igem.org/mediawiki/2010/7/74/BBF_RFC_64a.pdf]. | ||
| + | |||
| + | ===Activation experiments=== | ||
| + | |||
| + | [https://2010.igem.org/Team:Debrecen-Hungary/minimals#Cos-1_cells COS- 1 cells] were transfected with the chimeric receptors and reporter constructs. 5-6 hours after transfection we exposed the cells to ligand treatment (at least 10uM of ligands). | ||
| + | The CMX-GAL4-PPAR gamma was the positive control during our experiments, showing a stable and robust activation upon Rosiglitazone treatment, according to the b-galactosidase-normalized [https://2010.igem.org/Team:Debrecen-Hungary/minimals#Luciferase luciferase activity] levels. | ||
| + | |||
| + | We tested: | ||
| + | |||
| + | '''1)''' | ||
| + | |||
| + | ''pCDNA-GAL4-PXR-LBD | ||
| + | |||
| + | pSB1C3-GAL4-PXR-LBD'' | ||
| + | |||
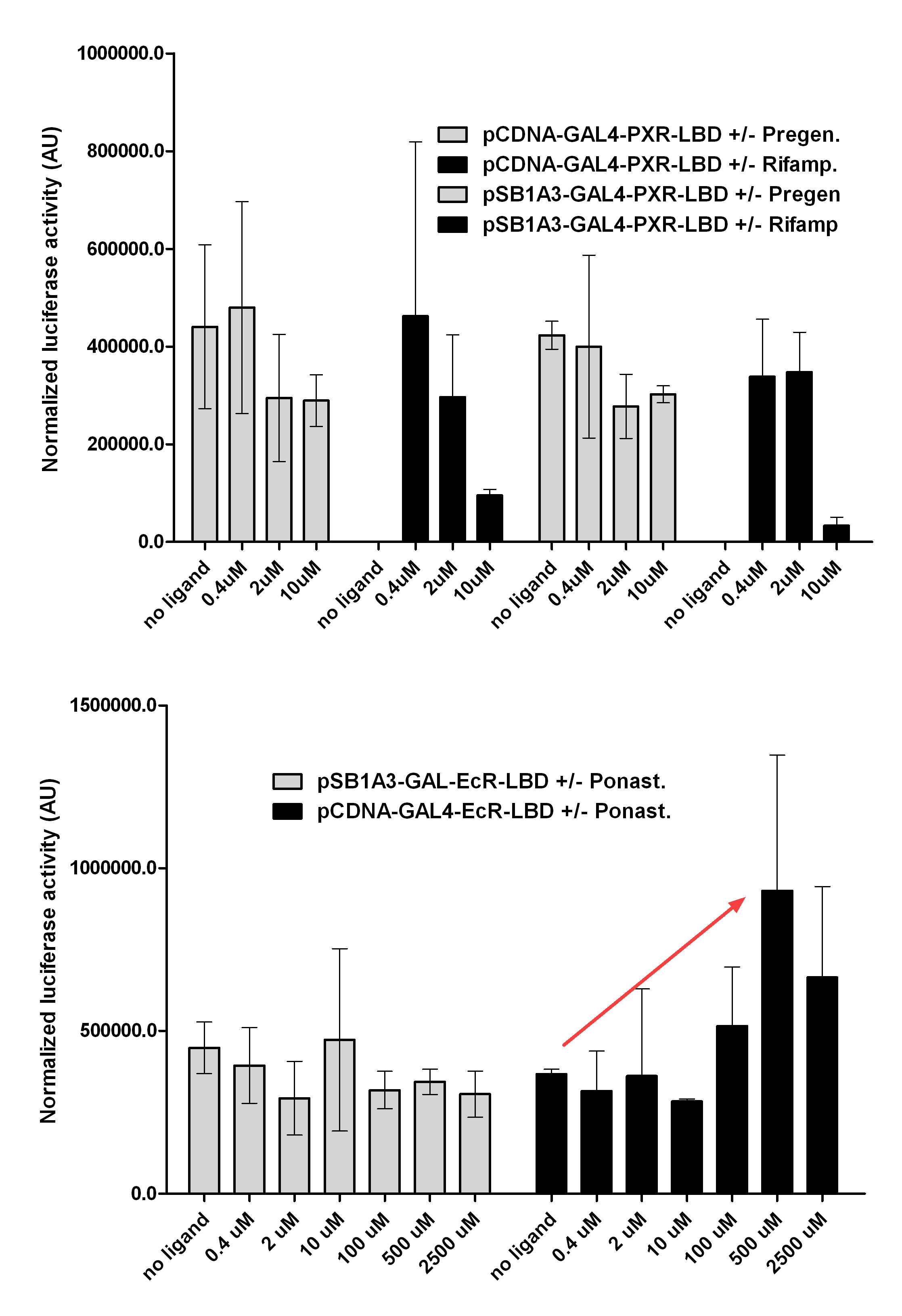
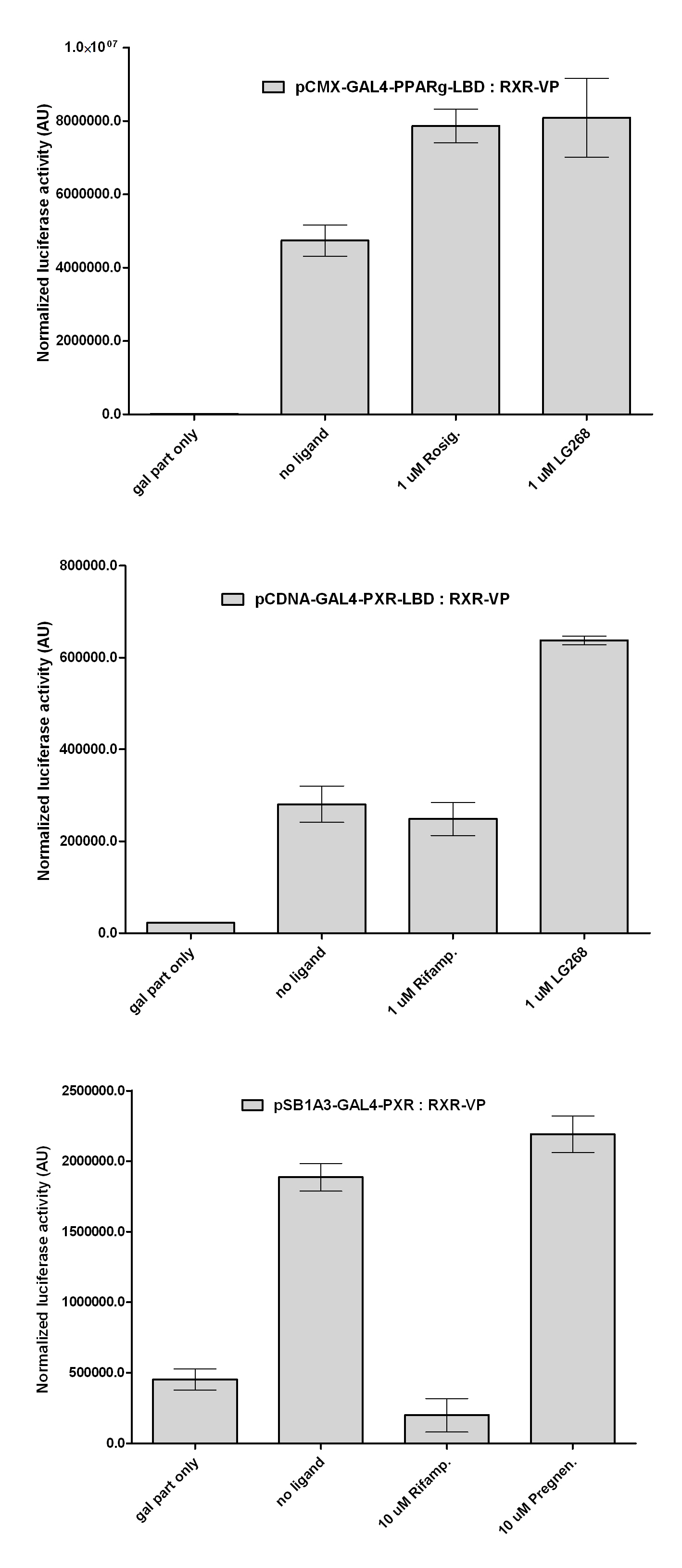
| + | with Pregnenolone carbonitrile, Rifampicin. These proteins showed no response to the mentioned ligands. | ||
| + | |||
| + | |||
| + | '''2)''' | ||
| + | |||
| + | ''pCDNA-GAL4-Ecdysone receptor-LBD | ||
| + | |||
| + | pSB1A3-GAL4-Ecdysone receptor-LBD'' | ||
| + | |||
| + | with Ponasterone A a typical ecdysteroid. Adding 5 uM of Ponasterone A resulted the activation of the construct (red arrow). | ||
| + | |||
| + | |||
| + | {| | ||
| + | | [[Image:PXR,_Ecr2.jpg|450px|thumb|upright|alt=|Test of pCDNA/pSB1A3-GAL4-PXR/EcR-LBD activity]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | ''pCDNA-GAL4-Estrogen receptor-LBD'' | ||
| + | |||
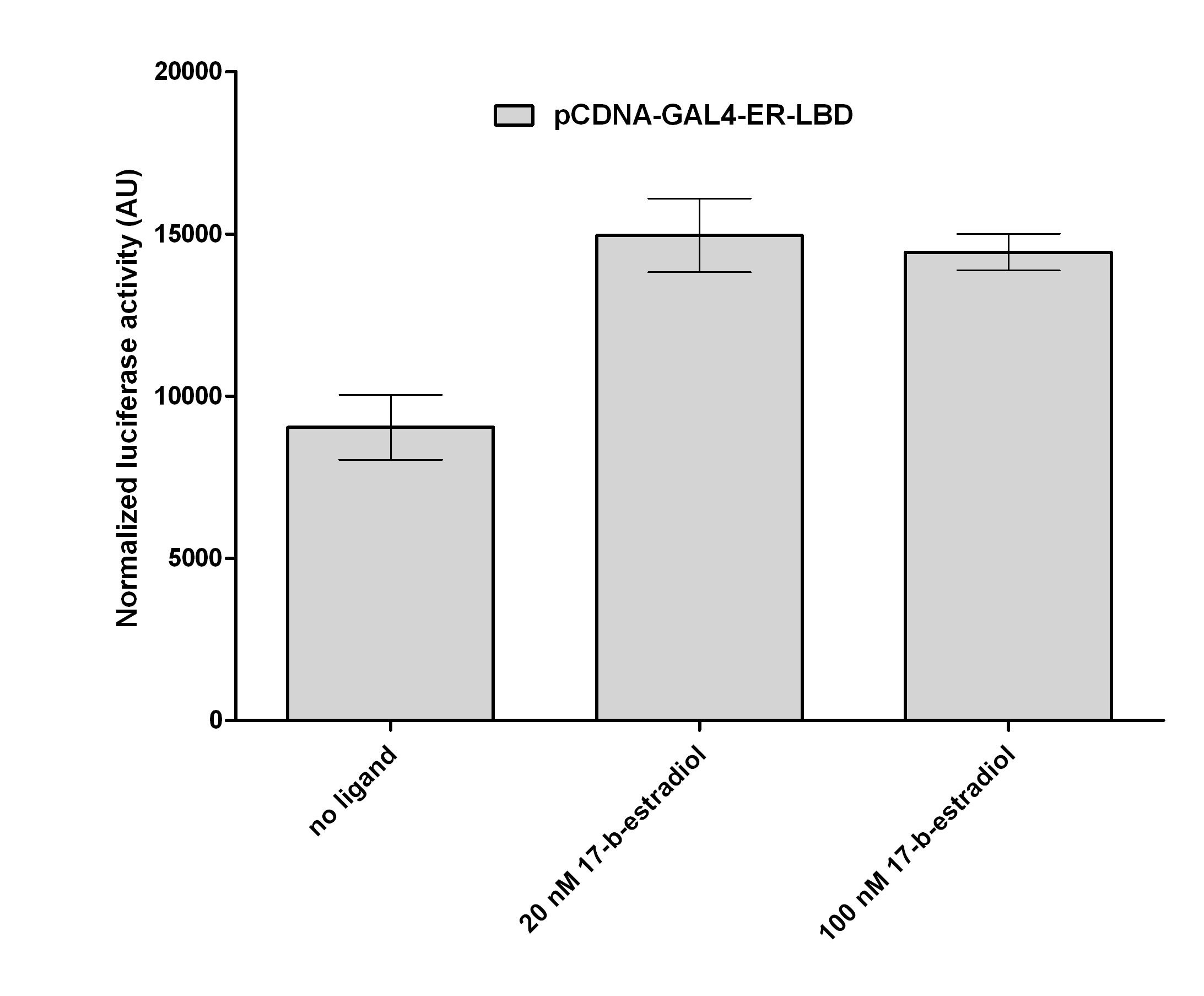
| + | with its known ligand, 17β-Estradiol. When we treated the cells with 100nM concentration of 17β-Estradiol, a 1.8 fold activation was detected, compare to the negative control (DMSO). | ||
| + | |||
| + | |||
| + | {| | ||
| + | | [[Image:ER2.jpg|450px|thumb|upright|alt=|Test of pCDNA-GAL4-ER-LBD activity]] | ||
| + | |} | ||
| + | |||
| + | ===Two hybrid system=== | ||
| + | |||
| + | Expression plasmids of the various receptor constructs were co-transfected into COS1 cells together with luciferase reporter plasmids. Our question was wether these proteins still have the '''ability to form RXR-heterodimers''', a key feature of several nuclear receptor superfamilies. In these experiments we used our newly fused parts and some characterized constructs that were partially '''published by our lab''': | ||
| + | |||
| + | CMX-VP-RXR-LBD | ||
| + | |||
| + | CMX-GAL4-PPAR gamma-LBD | ||
| + | |||
| + | CMX-GAL4-RAR alpha-LBD | ||
| + | |||
| + | CMX-GAL4-PXR-LBD | ||
| + | |||
| + | |||
| + | '''Our fused constructs:''' | ||
| + | |||
| + | |||
| + | pCDNA-GAL4-PXR-LBD | ||
| + | |||
| + | pSB1A3-GAL4-PXR-LBD | ||
| + | |||
| + | pCDNA-GAL4-Ecdysone receptor-LBD | ||
| + | |||
| + | |||
| + | As a positive control we transfected the cells with ''CMX-GAL4-PPARg-LBD or CMX-GAL4-PXR-LBD''. | ||
| + | Our results shows that the constructed chimeric recptors (''pCDNA-GAL4-PXR-LBD and pSB1A3-GAL4-PXR-LBD'') are '''able to interact''' with RXR-LBD. LG268 is a potent synthetic RXR-agonist, that enhances the formation of RXR-heterodimers. The increased luciferase expression of the last columns shows that dimers are formed. This is one key aspect of NR-action, that '''remained intact''' during our cloning process. | ||
| + | |||
| + | {| | ||
| + | | [[Image:Interact_3.jpg|450px|thumb|upright|alt=|Test of PXR:RXR interaction]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | <html><br><a name="plans"></a><br></html> | ||
| + | |||
| + | ='''BBF RFC 64 : Building Protein Domain Based Composite Biobricks for Mammalian Expression Systems'''= | ||
| + | The method described below is RECOMMENDED to be used for the design of mamalian domains that are planned to be used in mammalian expression vectors. It contains detailed instruction which may ensure protein expression of your transfected gene. [https://static.igem.org/mediawiki/2010/7/74/BBF_RFC_64a.pdf Click here to download the RFC] | ||
| + | |||
| + | ='''Plans, Possibilities'''= | ||
| + | |||
| + | The remotely activated transcription factor concept can be used to construct highly complex synthetic biological systems, for us it was a very appealing concept. Physicians may use fruit fly hormones in humans to titrate specific genes in gene therapy patient in selected tissue, such as dopamine receptor genes in schizophrenia. Worm species that can synthesize a different color reaction based on the amount of environmental pollutants in the soil they live. Scientists may induce stem cell pluripotency by titrating the exact amount of oncogenes needed for a fibroblast to turn into an embryo. | ||
| + | |||
| + | The defenite next step would be to add DBD's to the registry, or more exact a way for users to synthesize a DNA binding domain according to their sequance of interest. DNA binding domains are themselves modular from sybdomains as: helix loop helix, leucine zippers, helix turn helix and zinc fingers (the most popular subdomains). Once our team assembles a zinc finger toolkit with carefull construction instructions, a user could in vitro or in silico assemble and predict where in the genome his artificial transcription factor/ nuclear receptor will act on gene expression. In the future this will allow full freedom of harnesing the potency of artificial titratable transcription factors. | ||
| + | |||
| + | In our system we would like to induce [https://2010.igem.org/Team:Debrecen-Hungary/minimals#Apoptosis apoptosis] (click to find out more about apoptosis) by increasing the amount of proapoptotic proteins such as BAX by producing an expression vector containing TRE-minimal CMV promoter, BAX or PUMA and polyA tail. The pro-apoptotic proteins are able to express only in a controlled form using the Tetracyclin-controlled transcriptional activation system. The overexpression of these proteins can lead to Mitochondrial Outer Membrane Permeabilisation (MOMP) leading Cytochorome c release which is a point of no return in the mitochondrial pathway of Programmed Cell Death (PCD). The goal is to “switch off” the cells, which were genetically modified by us, after they “finished their work” in the recent local environment combining the components of the tetracycline-inducible system in self-contained retroviral and plasmid vectors. | ||
| + | |||
| + | Furthermore human, arthropode or C. elegans nuclear hormone receptor composite constructs can be expressed in an inducible form using [https://2010.igem.org/Team:Debrecen-Hungary/minimals#The_Tetracycline_On/Off_Gene_Expression_System Tetracyclin-controlled transcriptional activation] (click to find out more about TRE) system rather than using a vector which ensures constitutive expression of the parts. In our system Tetracycline Response Element (TRE) is recognized and bound by the Tetracycline repressor (TetR) protein. The TRE consists of 7 repeats separated by spacer sequences and placed upstream of CMV minimal promoter that has basal expession in the abscence of bound TetR. Tetracycline derivatives (e.g. doxycycline) bind TetR and render it incapable of binding to TRE, thereby forcing the expression of target genes. | ||
| + | |||
| + | |||
| + | <html><br><a name="contrib"></a><br></html> | ||
| + | |||
| + | ='''Contibution to project by team members'''= | ||
| + | |||
| + | *'''Students''' (in alphabetical order) | ||
| + | **'''Timea Beregi''' - Cloning work, wiki writing, team image, protocols writing | ||
| + | **'''Erika Berényi''' - Prepared psB1A3-TRECMV-polyA plasmid constructs( cloning, transfection, PCR), and prepared western blots after transfection. Uploaded description from apoptosis parts to the wiki and partsregistry. | ||
| + | **'''Shun Chieh Liu''' - Took part in the cloning task initially, demonstrated some in silico work, scriptwriter of the video projects, modified the side project page in the wiki. | ||
| + | **'''Lior Malka''' - Wiki design and programming, Culture team, Wiki Video Project design, Parts Registry moderator. | ||
| + | **'''Daniel Markovits''' - Wiki design and graphics, Help for teams PCR, Cell Culture and Luciferase teams, Video Project Narrator | ||
| + | **'''Katalin Nagy''' - Cloning work | ||
| + | **'''Lilla Ozgyin''' - Cloning work, coordinator of transfections, actress in video protocols, protocol writing, uploading a part, Parts Registry work, some wiki work, making photos of labwork. | ||
| + | **'''Katalin Sandor'''- Cloning, luciferase measurements, extraction of contaminated environmental samples, presenter of PCR video protocol, uploading to the wiki, making photos about labwork. | ||
| + | |||
| + | |||
| + | *'''Student Team''' Coordinators (in alphabetical order) | ||
| + | **'''Bence Daniel'''- Supervisor of the whole labwork, optimization of the luciferase measurements and the two hybrid system, 3D protein structures, wiki writing. | ||
| + | **'''Endre Kristof''' - Cloning (e.g. new expression vector from standard Biobrick parts), transfections and coordination in the wet lab. Participating in fund raising, in silico design, video and media project. Uploading materials of project description, future possibilities etc. to the wiki and informations about the composite and some basic parts to the partsregistry. | ||
| + | **'''Yakir Guri''' - Project selection and planning, part of cell culture team, cloning and in silico design. The supervisor of the [https://static.igem.org/mediawiki/2010/8/8f/Links_to_OWW_and_YOUTUBE.pdf 'Film in Lab'] video tutorial project (film making, editing, sound embedding) and [http://www.youtube.com/user/debrecenigem2010 Youtube channel] manager. Collecting, editing and reviewing protocols and uploading to the [http://www.openwetware.org/wiki/Special:Contributions/Yakir_Guri 'Open Wet Ware'] . Coordinator of PR and fundraising (media, images, writing [https://static.igem.org/mediawiki/2010/2/2a/Medikuslap.pdf articles], interviewing team members). | ||
| + | **'''Ophir Keret''' - Intiated project together with Balint.L.Balint, Team recruitment, project selection, work planning and coordination, team tutorials, labwork supervision, in silico design/modeling of the parts, construction of the expression vector, PCR work, Chief wiki editor, contributed to wiki writings (abstract, introduction, minimals, on the media). | ||
| + | |||
| + | |||
| + | *'''Team Instructors'''(in alphabetical order) | ||
| + | **'''Balint L. Balint''' - Initiated the project, made the team recruitment, fund raising, some of the PR, laboratory setup, helped shaping the project, seting up of the team. I supervised the first lab experiments and planned some of the experiments later on, ordered parts and reagents, organized finances, organized administrative issues of the trip. Supervised wiki and partsregistry work. | ||
| + | **'''Peter Brazda''' - Supervised planing and evaluation of luciferase assays. Participated in wet-lab education and the wiki-works. Co-mentored students in discussing scientific problems and questions of NR-biology. | ||
| + | **'''Mate Demeny''' - Implementation of the general approach for the design of the BBF_RFC 25 compatible parts. Supervised the cloning process and designed adaptors to the pCDNA3.1. Tutoring the cloning process. Troubleshooting for the cloning team. | ||
| + | **'''Gabor Zahuczky''' - Supported the initiation of the project, fund raising, organization of finances and administration. Participated in the design of parts and wet lab education. | ||
| + | <html> | ||
<center> | <center> | ||
| - | <a href="https://2010.igem.org/Team:Debrecen-Hungary"><img src=" | + | <a href="https://2010.igem.org/Team:Debrecen-Hungary"><img src="https://static.igem.org/mediawiki/2010/9/96/Backbuttonbck.jpg"></a> |
</center> | </center> | ||
| - | |||
</html> | </html> | ||
Latest revision as of 19:16, 27 October 2010
|
AbstractEukaryotic synthetic biology has huge potential, yet it is still in need of more diverse molecular tools for defined gene regulation. Nuclear receptors are a conserved family of proteins responsible for sensing lipids; they may be viewed as lipid activated transcription factors. We have successfully developed a kit with a variety of lipid responsive domains (from H.sapiens, D.melanogaster and C.elegans) for the rational construction of synthetic transcription factors. The domains respond only to predefined lipids and selectively activate predetermined gene expression. To characterize theses domains, we used standardized protocols for comparable measurements. In vivo gene expression was measured as a function of ligand concentration using luciferase activity.
IntroductionSynthetic BiologySynthetic biology is a relatively new area of biological research; similar to many other new scientific fields it has many definitions. The best way to express the meaning of synthetic biology is to understand the desired end-result: engineering of complex biological systems. These systems are best thought of as analogues of everyday machinery: cogwheels, levers, timers, button and buzzers (in this case a clock), only in the case of biological systems (molecular ones) cells, DNA, proteins, lipids, sugars and RNA are the “parts” of the system. Similar to mechanical engineering (or every other engineering branch) there is a need for standards (consensus way of doing things), abstraction (simple and unified way of thinking about the parts of a system), and modularity (how these parts interact to become the device, or several devices into a system). Thus a good definition for synthetic biology could be engineering of molecular (for the time being) biological systems according to preset standard parts. The international genetically engineered machines and associated parts registry are, to date, one of the largest registries for standard parts in use for synthetic biology. Free information can be found in the registry regarding parts, devices and modules all inputted by various teams worldwide. Eukaryotic synthetic biology is still in its infancy. The large kingdom of metazoans includes all multicellular eukaryotes such as mammalians, arthropods and nematodes. No standard chassis (framework) exists for the animal kingdom which makes them far less popular then the famous E.coli. The protein modules derived from metazoans (like Drosophila or C. elegans) are functional in yeasts also. Very few iGEM teams (or even labs outside iGEM) have chosen to toggle the animal chassis (two notable examples are team Heidelberg 2009 and team Slovenia 2006). The amount of available compatible parts is limited, which severely restricts the options of creating complex biological devices. Nearly no imagination is required for designing tools, since their analogues already exist in the bacterial chassis. The possible use of such systems is unlimited. Field’s such as of environment, medicine, energy and research all gain to profit from the development of animal synthetic biology. Systems requiring gene expression input in eukaryotic synthetic biology systems require a way to standardize gene expression, a complicated task. The way from gene to protein contains many steps of possible error: transcription factor binding, promoter strength, recruitment of auxiliary proteins, nuclear RNA synthesis and many more steps finally leading to translation, folding, cleaving and delivery (but hey, you have to start somewhere). Our team was interested at designing eukaryotic synthetic biology tools related to PoPs. PoPs (polymerase per second), the flying Dutchman of synthetic biology, is a number which represents the rate (base pair per second) at which RNA polymerase crosses past a given DNA position. Currently, no in vivo technique for measuring PoPS directly exists; it can be estimated indirectly by measuring other parameters (eg protein expression or enzyme activity). Nevertheless it is still a useful abstraction for thinking about transcription-based logic devices and it allows the engineer to define devices. Our aim, was not only to infer PoPs but to devise a way to titrate it remotely.
New Tools for Synthetic Biology: Nuclear ReceptorsNuclear receptors (click to find out about nuclear receptors), best viewed as transcription factors which can be activated by extracellular cues, are unique in their ability to allow direct remote PoPs titration (both activation and repression). These receptor classes bears high homology to each other throughout the animal kingdom and are modular into distinct domains. All of these features attracted our attention to find a way to incorporate these tools in the parts registry, and characterize them in standardized methods. Many nuclear receptors, are among the primary targets of drug discovery because of their diverse biological actions. Nuclear receptors bear high homology to each other and are modular into distinct domains: N-terminal regulatory domain, DNA-binding domain (DBD), a Hinge region, Ligand binding domain (LBD) and a C-terminal domain. The LBD (click to find out more about LBD's) is the segment of the receptor which changes its conformation upon lipophilic ligand binding; this also causes the exposure of a powerful transcriptional activation domain (which attracts the transcriptional machinery). This was our segment of interest since it links ligand binding with transcriptional activity. Our team has generated a library of ligand binding domains for the rational construction of synthetic titrateable transcription factors. In our model hybrid receptor, this LBD was fused to a Gal4 DNA binding domain. The sequence to which Gal4 binds is CGG-N11-CCG, where N can be any base. Although Gal4 is a yeast protein not normally present in other organisms it has been shown to work as a transcription factor in a variety of organisms such as Drosophila, and human cells, highlighting that the same mechanisms for gene expression have been conserved over the course of evolution. The examination of these chimeric receptors was through a version of the two hybrid assay (click to find out more about the two hybrid assay). For a more detailed description of our experimental system, please visit our "Minimals" page. ResultsFirst we had to generate expression vectors that can be used in mammalian expression systems. The cloning work of Team Debrecen-Hungary resulted 18 basic parts in pSB1C3 and 13 composite parts in pSB1C3 or pSB1A3. All of them have been tested by restriction enzyme analysis and by sequencing. Furthermore, some basic and fused parts have been ligated into pCDNA3.1 containing a specific adaptor by which RFC-25 compatible parts can be inserted into this expression vector. pCDNA3.1 results constitutive expression of the inserted part in the cells so the investigated part can be tested at functional level. The other expression vector is a fully SB (Synthetic Biology) compatible mammalian expression vector generated from pSB1A3. We inserted into the pSB1A3 the TRE (tetracycline response element) a strong CMV (cytomegalo virus) promoter and a PolyA element that is a terminator of transcription in mammalian cells. These four elements are available as individual parts in the parts registry and can be used in combination with other parts too. CloningBasic Parts18 basic parts have been inserted into pSB1C3 which contain 3 DBDs, 1 hinge, 11 LBDs, 2 regulators and 1 apoptosis protein. A Gal4 DBD which has the RFC25 N terminal linkers was designed and submitted to be used (BBa_K364319). Composite Parts13 composite parts have been created, ten of them consist of two basic parts, one of them has been built up from three basic parts, and an expression vector from standard biobrick parts has been created by using four basic parts. The backbone has been opened with AgeI and PstI restriction enzymes in order to ligate the part cut out with NgoMIV and PstI. The previous restriction digests have been repeated so each part could be inserted after the previous one. Gal4-human LBD fusionsTwo human nuclear hormone receptor LBDs (Pregnane X Receptor LBD, Estrogen Receptor LBD) have been inserted into pSB1C3 after Gal4 was ligated into the backbone. The Gal4 (438 bp) is a modular protein consisting of a DNA-binding domain and an activation domain which can be used in Two-Hybrid system in order to identify interactions between two proteins or a protein with DNA. The length of PXR LBD is 732 bp while the ER LBD is 828 bp long, so the fusion leads to 1170 bp and 1266 bp long composite inserts. Both of them have been transferred into pCDNA3.1 to express them in certain cell lines. Gal4-Ecdysone receptor fusionThe LBD of Ecdysone Receptor (639 bp) which can be found in arthropodes has been inserted after the Gal4 both in the pSB1C3 and pCDNA3.1 resulting an 1077 bp long insert in the vectors. Gal4-C. elegans LBD fusionsFive nuclear hormone receptor LBDs of C. elegans (NHR8, NHR23, NHR31, NHR80, DAF12) have been inserted after Gal4 in pSB1C3, resulting inserts with the lenght between 944 bp and 977 bp. All of the Gal4-C.elegans LBD constructs (e.g. Gal4-NHR8) except Gal4-NHR80 composite part have been fused into pCDNA3.1 expression vector to test them as a lipid sensor. Estrogen Receptor from basic partsEstrogen receptor DBD (318 bp) and LBD (828 bp) have been inserted together into pSB1C3. The two main domains of ER are linked together by the hinge region of Vitamin D Receptor (357 bp) resulting a 1503 bp long insert in the backbone. Creating expression vector from pSB1A3Three basic parts from pSB1C3 have been ligated into pSB1A3 consecutively by using chloramphenicol and ampicillin assembly in order to create an expression vector from the ampicillin resistance carrying backbone. The first step was to insert 321 bp long TRE-CMV (Tetracyclin Response Element- Minimal Cytomegalovirus promoter) into pSB1A3 which has the length of 2157 bp. Then Gal4-PXR LBD (1170 bp) and Gal4-Ecr LBD (1077 bp) fusions were inserted after the TRE- CMV, before PolyA tail (191 bp) was ligated in the end.
Western Blots of Gal-fused proteinsThe functionality of the constructs was assesed by Gal4 Western Blot. 293T cells were transfected. Western-blot was carried out using rabbit anti-Gal4 polyclonal primary antibody. High protein expression have been noticed when Gal4-PXR LBD composite construct was cloned into pCDNA3.1 as well as into the expression vector created from pSB1A3 (using standard BioBrick parts). Lower expression was detected in the case of Gal4-EcR LBD in both expression vectors. The Gal4-PXR and EcR-LBD fused proteins appeared at about 40 kDa while the Gal4-C.elegans-LBD (NHR8, NHR31) chimeric proteins which have been intensely expressed, can be seen between 30-35 kDa which corresponds to thier predicted molecular weight. Transients cotransfection assay /two-hybrid system/We moved the composite parts generated in pSB1C3 into the expression vectors, then transfered these fused parts into pCDNA and pSB1A3 vectors for further works. pCDNA is designed for high-level expression, purification and detection of recombinant protein in mammalian hosts. In addition we developed a new expression vector pSB1A3 with a CMV-promoter and poly-A tail. Our experiments proceeded on two different lines. The first one is the presentation of the cloned constructs bearing the ability to get activated by ligand binding. These were the activation experiments. Furthermore we showed that our constructs had the ability to interact with the previously published heterodimeric partner RXR. To confirm this we used the mammalian two-hybrid system. Binding to RXR confirms the proper folding and ultimately the generation of a well designed LBD, and a functional mammalian expression vector. The method used to design our parts is described as a proposed standard BBF RFC 64 [1]. Activation experimentsCOS- 1 cells were transfected with the chimeric receptors and reporter constructs. 5-6 hours after transfection we exposed the cells to ligand treatment (at least 10uM of ligands). The CMX-GAL4-PPAR gamma was the positive control during our experiments, showing a stable and robust activation upon Rosiglitazone treatment, according to the b-galactosidase-normalized luciferase activity levels. We tested: 1) pCDNA-GAL4-PXR-LBD pSB1C3-GAL4-PXR-LBD with Pregnenolone carbonitrile, Rifampicin. These proteins showed no response to the mentioned ligands.
pCDNA-GAL4-Ecdysone receptor-LBD pSB1A3-GAL4-Ecdysone receptor-LBD with Ponasterone A a typical ecdysteroid. Adding 5 uM of Ponasterone A resulted the activation of the construct (red arrow).
pCDNA-GAL4-Estrogen receptor-LBD with its known ligand, 17β-Estradiol. When we treated the cells with 100nM concentration of 17β-Estradiol, a 1.8 fold activation was detected, compare to the negative control (DMSO).
Two hybrid systemExpression plasmids of the various receptor constructs were co-transfected into COS1 cells together with luciferase reporter plasmids. Our question was wether these proteins still have the ability to form RXR-heterodimers, a key feature of several nuclear receptor superfamilies. In these experiments we used our newly fused parts and some characterized constructs that were partially published by our lab: CMX-VP-RXR-LBD CMX-GAL4-PPAR gamma-LBD CMX-GAL4-RAR alpha-LBD CMX-GAL4-PXR-LBD
pSB1A3-GAL4-PXR-LBD pCDNA-GAL4-Ecdysone receptor-LBD
BBF RFC 64 : Building Protein Domain Based Composite Biobricks for Mammalian Expression SystemsThe method described below is RECOMMENDED to be used for the design of mamalian domains that are planned to be used in mammalian expression vectors. It contains detailed instruction which may ensure protein expression of your transfected gene. Click here to download the RFC Plans, PossibilitiesThe remotely activated transcription factor concept can be used to construct highly complex synthetic biological systems, for us it was a very appealing concept. Physicians may use fruit fly hormones in humans to titrate specific genes in gene therapy patient in selected tissue, such as dopamine receptor genes in schizophrenia. Worm species that can synthesize a different color reaction based on the amount of environmental pollutants in the soil they live. Scientists may induce stem cell pluripotency by titrating the exact amount of oncogenes needed for a fibroblast to turn into an embryo. The defenite next step would be to add DBD's to the registry, or more exact a way for users to synthesize a DNA binding domain according to their sequance of interest. DNA binding domains are themselves modular from sybdomains as: helix loop helix, leucine zippers, helix turn helix and zinc fingers (the most popular subdomains). Once our team assembles a zinc finger toolkit with carefull construction instructions, a user could in vitro or in silico assemble and predict where in the genome his artificial transcription factor/ nuclear receptor will act on gene expression. In the future this will allow full freedom of harnesing the potency of artificial titratable transcription factors. In our system we would like to induce apoptosis (click to find out more about apoptosis) by increasing the amount of proapoptotic proteins such as BAX by producing an expression vector containing TRE-minimal CMV promoter, BAX or PUMA and polyA tail. The pro-apoptotic proteins are able to express only in a controlled form using the Tetracyclin-controlled transcriptional activation system. The overexpression of these proteins can lead to Mitochondrial Outer Membrane Permeabilisation (MOMP) leading Cytochorome c release which is a point of no return in the mitochondrial pathway of Programmed Cell Death (PCD). The goal is to “switch off” the cells, which were genetically modified by us, after they “finished their work” in the recent local environment combining the components of the tetracycline-inducible system in self-contained retroviral and plasmid vectors. Furthermore human, arthropode or C. elegans nuclear hormone receptor composite constructs can be expressed in an inducible form using Tetracyclin-controlled transcriptional activation (click to find out more about TRE) system rather than using a vector which ensures constitutive expression of the parts. In our system Tetracycline Response Element (TRE) is recognized and bound by the Tetracycline repressor (TetR) protein. The TRE consists of 7 repeats separated by spacer sequences and placed upstream of CMV minimal promoter that has basal expession in the abscence of bound TetR. Tetracycline derivatives (e.g. doxycycline) bind TetR and render it incapable of binding to TRE, thereby forcing the expression of target genes. Contibution to project by team members

|
 "
"
 Team
Team