Team:UIUC-Illinois-Software/Project
From 2010.igem.org
 | ||
Contents |
[http://igem.illinois.edu/ BioMORTAR website]
BioMORTAR Project Abstract
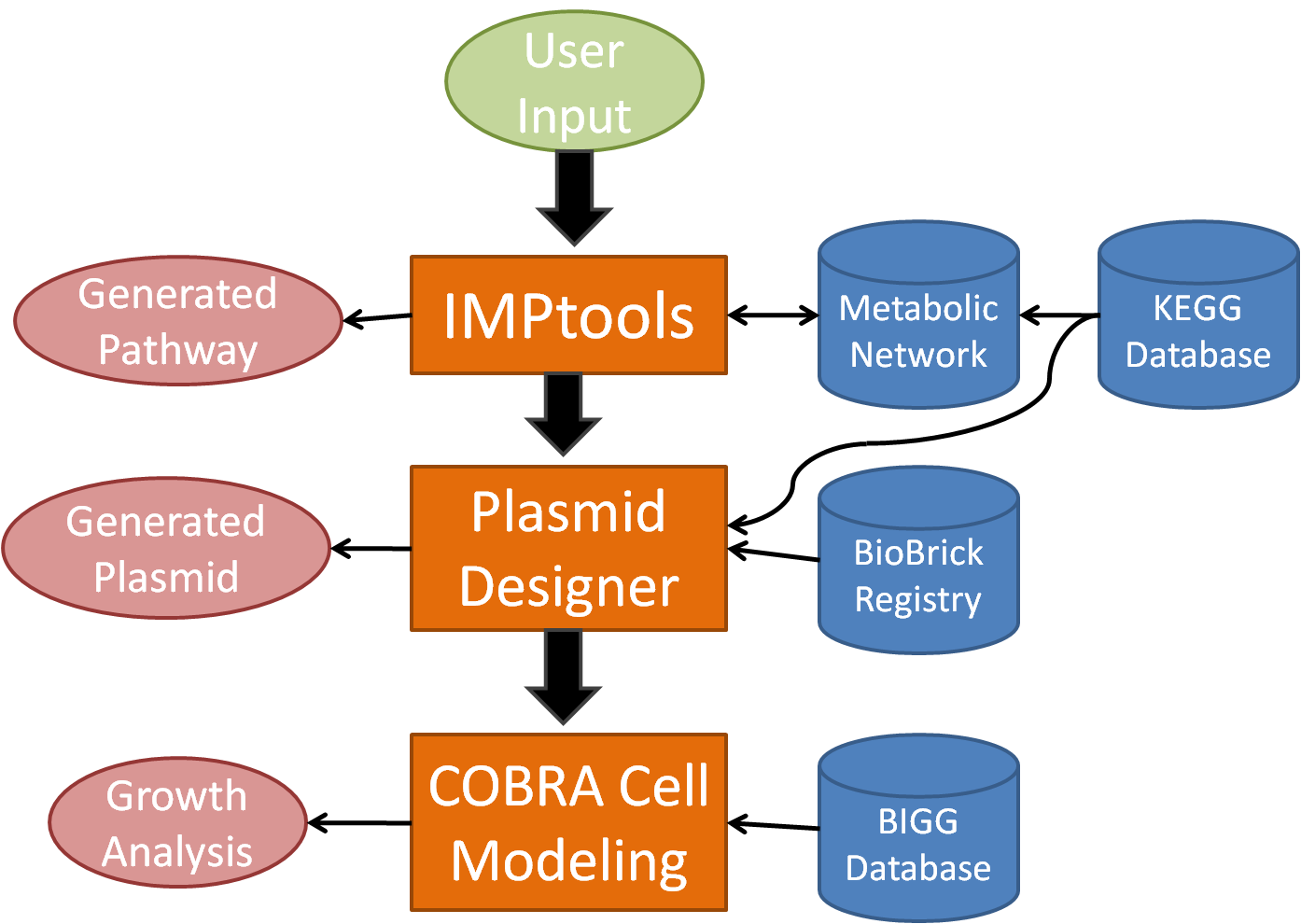
In order to facilitate the design process for novel bacterial metabolism, our team has created a tool suite known as BioMortar which will automate plasmid design for metabolic processes as well as model cell growth. BioMortar begins with a much improved version of IMPtools, which uses an algorithm over a network generated by the KEGG database, to determine the optimal metabolic pathway according to specified conditions. At this point, it accesses the DNA sequences for each recommended enzyme for each reaction and searches the BioBrick database for related gene sequences. Then, BioMORTAR designs and displays the advised, usable plasmid(s) in BioBrick format for the user. Finally, the program models the growth of the organism, with the addition of the new metabolic pathway(s). By automating the design process, BioMORTAR streamlines the process of designing bacteria with new metabolic processes.
Project Description
The 2010 Illinois iGEM Tools Team is in the phase of constructing a program to design a plasmid in-silico for the user-specified host bacteria as well as model the growth of the designed cell.
The web-based program will begin by implementing the desired metabolic pathway computed by a greatly improved version of last year's project, IMPtools, then by determining the recommended enzymes for each reaction, and accessing the DNA sequence for it. In the meantime, the program will model the growth of the user specified bacteria with the new metabolic pathway to optimize the plasmid design. Finally, it will construct and display, with annotation, the advised plasmid(s) for the user as well as display important information on projected growth. In addition, it will have the option to adhere to the iGEM biobrick standards and will reference the biobrick database.
As a whole, the program will not only give the user the optimal plasmid design for the metabolic process, but it will project growth for the modified bacteria and connect the user with the BioBrick database which can reference the necessary plasmids.
Purpose/Application
The BioMORTAR project was motivated by the need for a systems approach to assembling components in synthetic biology, and to expedite the design process in metabolic engineering. Since the science of synthetic biology is becoming compartmentalized, the design process should keep up with this progress, and allow for a quick and simple way to find genetic parts for a specific purpose, connect the parts, then simulate a novel cell in silico.
One possible application of BioMORTAR is to use the program to quickly design a library of bacteria to combat an array of possible chemical or biological threats. Other applications for this program are focused towards, but not limited to, the design of large scale metabolic processing and any other function requiring the use of novel bacteria cells to carry out engineered metabolic reactions.
BioMORTAR 3-Part Design Suite
Progression of BioMORTAR
IMPtools - The first part of BioMORTAR is IMPtools, a metabolic pathway generator that allows the user to input an input and output compound, then traverse a reaction network, built from the KEGG Database, to generate the optimal metabolic pathway.
Plasmid Designer - The second part of BioMORTAR aggregates the coding DNA for the enzymes used in the metabolic reactions generated in IMPtools, then joins the DNA sections with user selected promoters, ribosome binding sites, and terminators. The plasmid designer attaches it all to a vector to create the plasmid that would allow a cell to carry out the generated metabolic process. The plasmid designer then searches the BioBrick iGEM parts registry for similar sequences within the plasmid, and references these existing parts to the user.
COBRA Cell Modeling - The third and final part of BioMORTAR is cell modeling using the BIGG database in conjunction with COBRA tools in MATLAB to simulate cell growth and chemical flux from the added pathway within the cell.
IMPtools (AKA IMPtools 2.0)
The first release of IMPtools is the result of the efforts of the 2009 University of Illinois iGEM software team. The web-based program works by finding the quickest pathway between two organic compounds over a reaction network constructed from the KEGG database. It also would allow the user to customize the algorithm by using a weighting system to specify the importance of certain features of reactions, such as ATP consumption, metabolite connectivity, and amount of extra product at the end. The software won best software as well as a gold medal in the 2009 iGEM competition at MIT, however it still had a few problems to work out before it could be feasibly used.
IMPtools has had some big improvements made to it since last year's iGEM competition. First, some major bugs in the program were fixed, and the inputs were simplified to recognize the name of the compound rather than merely the KEGG ID. There are two major improvements to IMPtools. The first and most fundamental, is the reduction of the rather bloated network by the application of the RPAIR database available from the KEGG project. The RPAIR database pairs compounds that chemically interact for each reaction, making the network a much smarter network to work with. The new IMPtools relies on the direct nature of the links for greater accuracy in generating a realistic metabolic pathway. Another large improvement is the addition of a branching system. One major drawback of IMPtools was that the algorithm would simply ignore large intermediate inputs and outputs in the pathway. The branching system solves this by keeping track of these large side products and attempting to convert them into readily available reactants or common metabolic products that could be provided or absorbed by the cell's existing metabolism.
- User inputs the desired starting and ending compounds
- Weighting system allows for a more customized metabolic pathway
- IMPtools selects the best metabolic pathway from the generated reaction network
- An option allows IMPtools to optimize output while minimizing intermediate inputs or products through pathway branching
Plasmid Designer
Building on top of IMPTools, BioMORTAR plasmid designer determines the requisite enzymes and retrieves the appropriate gene sequences from the KEGG database. BioMORTAR will then assemble a plasmid or an array of plasmids in silico for the necessary metabolism according to the expression characteristics designated by the user. The target organism is accounted for when collecting the necessary genetic code - as it may have already have the metabolic capability coded into its genome. Additionally, BioMORTAR will consider the existing biological library, the BioBrick registry, which is submitted to and verified by the Registry of Standard Biological Parts. BioMORTAR plasmid designer then finds an enzyme coding BioBrick sequence in the registry and uses it in the plasmid as appropriate. BioMORTAR also provides an easy-to-use interface for selecting the regulatory BioBrick parts for highly tuned pathway expression, including a given plasmid backbone, promoter sequence, ribosome binding site , and terminator sequence. Such controls allow for the novel pathway to be constitutive or regulated, the backbone plasmid to be multiplicative in the host (high copy number), or for complex self-regulation of the novel pathway (feedback inhibition).
- Reads the enzymes from the generated metabolic pathway
- Searches the databases for the genes for the required enzymes
- Attaches specified promoters, ribosome binding sites, and terminators
- Organizes all of the genetic code and creates a plasmid in silico
- References relevant genes on various databases, allowing the user to easily create the plasmid in the lab
COBRA cell Modeling
The final part of BioMORTAR is the in silco prediction of cellular behavior and growth with the incorporation of the plasmid into a selection of organisms, more specifically Escherichia coli iJR904, iAF12 and the textbook strains; Helicobacter pylori iIT341; Methanosarcina barkeri iAF692; Mycobacterium tuberculosis iNJ661; Staphylococcus aureus iSB619; Saccharomyces cerevisiae iND750. The genome-scale metabolic reactions and pathway reconstruction models have been provided by the Biochemical, Genetic and Genomic knowledge base of large scale metabolic reconstructions, or BiGG (Schellenberger, 2010) in the Systems Biology Markup Language (SBML) format. To assemble predicted growth and cellular behavior, the model is loaded into the Constraint-Based Reconstruction and Analysis Toolbox (COBRA) using the Matlab software (Becker, 2007). This program is able to construct reaction fluxes of a biochemical reaction based on user-defined bounds for fluxes through reactions within the metabolic network. The COBRA program uses dynamic flux-balance analysis (FBA) to predict the maximum growth rates given in biomass composition. The flux variability analysis (FVA) will provide reaction flux within a network, specifically through the reactions of interest.
- Calculates the growth rate and robustness of bacteria with the added metabolic pathway
- Determines the feasibility of survival with the new reactions
- May be able to optimize the plasmid for survivability
Program Design
BioMORTAR is web-based, and can be found linked to by our iGEM wiki page. BioMORTAR uses different coding languages to perform different functions more efficiently. IMPtools and the plasmid designer are coded in Python since the language is so versatile, whereas the cell modeling part of BioMORTAR is coded in MATLAB since COBRA provides many of the tools necessary to model cell performance. The user interface for all of BioMORTAR is programmed in Django, a high level Python web framework that allows for a simple user interface, in our case, and a simple setup for hosting the program on our server. The server is owned and operated by the UIUC iGEM team, and it currently runs from its location at the University of Illinois.
Judging Criteria
The requirements to earn a Bronze Mousepad are:
- Register the team, have a great summer, and have fun attending the Jamboree.
- Create and share a Description of the team's project via the iGEM wiki.
- Present a Poster and Talk at the iGEM Jamboree.
- Develop and make available via the Registry an open source software tool that supports synthetic biology based on BioBrick standard biological parts (remember, the iGEM judges will be looking for substantial team-based software projects).
The requirements to earn a Silver Mousepad, in addition to the Bronze Mousepad requirements, are:
- Provide a detailed, draft specification for the next version of your software tool, or a second, distinct software tools project.
The requirements to earn a Gold Mousepad, in addition to the Silver Mousepad requirements, are:
- Help another iGEM team by, for example, analyzing a Part, debugging a Device, or modeling or simulating a System.
- Develop and document a new technical standard that supports the (i) design of BioBrick Parts or Devices, or (ii) construction of BioBrick Parts or Devices, or (iii) characterization of BioBrick Parts or Devices, or (iv) analysis, modeling, and simulation of BioBrick Parts or Devices, or (v) sharing of BioBrick Parts or Devices, either via physical DNA or as information via the internet.
- Outline and detail a new approach to an issue of Human Practice in synthetic biology as it relates to your project, such as safety, security, ethics, or ownership, sharing, and innovation.
References
Schellenberger, J., Park, J. O., Conrad, T. C., and Palsson, B. Ø., "BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions", BMC Bioinformatics, 11:213, (2010).
Becker, S.A., Feist, A.M., Mo, M.L., Hannum, G., Palsson, B. Ø., Herrgard, M.J., “Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox”, Nature Protocols¸2:3:727-738, (2007).
 "
"