Team:TU Delft/Project/rbs-characterization/characterization
From 2010.igem.org
(New page: =Characterization= <html><div style='clear:both'> The cells were cultured over 18 hours in 96-well plates using a Gen5 fluorescence and absorbance plate reader, LB with ampicillin was the ...)
Newer edit →
Revision as of 16:58, 10 September 2010
Characterization
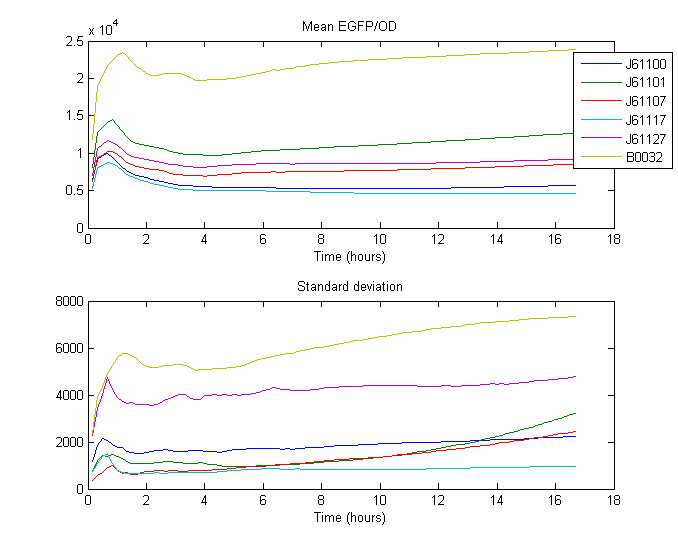
Note: Fluorescence (y-axis) is reported as Arbitrary Fluorescence Units.
The RBS strength was calculated by taking the mean of the ratio between the expression of the standard RBS ([http://partsregistry.org/Part:BBa_B0032 B0032]) and expression of Anderson RBS over time. The Expression is defined as the quotient of the measured fluorescence divided by measured biomass (OD at 600nm).
| RBS | Strength |
| [http://partsregistry.org/Part:BBa_J61100 J61100] | 0.047513 |
| [http://partsregistry.org/Part:BBa_J61101 J61101] | 0.119831 |
| [http://partsregistry.org/Part:BBa_J61107 J61107] | 0.065454 |
| [http://partsregistry.org/Part:BBa_J61117 J61117] | 0.038518 |
| [http://partsregistry.org/Part:BBa_J61127 J61127] | 0.087334 |
| [http://partsregistry.org/Part:BBa_B0032 B0032] | 0.300000 |
The result of this experimental set up was a simple characterization of five of the Anderson RBS sequences in relation to other standard well-characterized RBS. The given relative strengths are displayed assuming [http://partsregistry.org/Part:BBa_B0034 B0034] as the unit.
We're also trying to look into mRNA folded shapes using mfold to see if there is a common pattern in the Anderson RBS shapes. This might be usable in predicting RBS strength for the other untested Anderson RBS sequences. Unfortunately, this seems not to be the case, as all the RBS sequences have very different mfold shapes.
Source data
Source code and data used for characterization:
Data: Experiment1_26_7_10.m
Matlab calculation code: IGEM_TUDelft_2010_Rbs_calc.m
 "
"