Team:St Andrews/project/laboratory
From 2010.igem.org
(→The CAI-1 sender) |
(→The CAI-1 sender) |
||
| (40 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
== Overview == | == Overview == | ||
| - | + | This part of the lab work has focused on constructing two main biobricks - the bistable switch and the CAI-1 sender. | |
===The bistable switch=== | ===The bistable switch=== | ||
| + | To acheive bistable expression patterns in quorum sensing systems, we planned to rearrange the Lux operon so that both LuxI and LuxR were downstream of the Lux promotor. | ||
| - | + | [[Image:LuxOpWiki.png|600px]] | |
| - | |||
| - | |||
| - | + | To do this we planned to use already available parts from the registry to assemble our bistable operon. | |
| - | + | [[Image:BuildingOperon.png|600px]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | We encountered problems with both 3 antibiotic assembly and In-Fusion PCR based assembly. These are detailed in the lab nootbook, but we eventually decided we had spent far too much time debugging our assembly methods and needed to move onto other areas of the project. The bistable operon was never finished. | |
| - | === | + | ===Reference=== |
| - | + | EL Haseltine, FH Arnold (2007) - Implications of Rewiring Bacterial Quorum Sensing. Applied and Environmental Microbiology 74-2 437-445 | |
| - | + | ===[http://partsregistry.org/Part:BBa_K356000 The CAI-1 sender]=== | |
| - | + | This biobrick is essential to our project. CAI-1, the cholera-specific auto inducing molecule, can switch off the production of cholera toxin by ''Vibrio cholerae'' when it is present in high concentrations. To make this biobrick the gene responsible for making the CAI-1 synthase (CqsA) was optimised for E. coli and then synthesised by geneart. | |
| + | CqsA can be found [http://partsregistry.org/Part:BBa_K356001 here]. | ||
| - | First | + | First we found a sequence for the cholera gene for ''Vibrio cholerae'' autoinducer synthase. |
| - | + | [http://www.genome.jp/dbget-bin/www_bget?vch:VCA0523 the CAI-1 autoinducer synthase at genome.jp] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | + | We then codon optomised this gene for ''Escherichia coli'' using the functionality built into [http://mrgene.com/desktopdefault.aspx/tabid-2/ the Mr Gene Page] | |
| - | + | which gave us | |
| - | + | <p>ATGAACAAACCTCAGCTGCCTGACTTTATCCAAAACAAAATCGACCACTATATCGAGAACT | |
| + | ATTTCGACATTAACAAAAACGGCAAACACCTGGTGCTGGGCAAACAAGCATCACCGGATGA | ||
| + | CATTATCCTGCAAAGCAACGACTATCTGGCCCTGGCTAATCACCCTCTGATCAAAGCTCGT | ||
| + | CTGGCGAAAAGCCTGCTGGAAGAACAGCAATCCCTGTTTATGAGCGCCTCCTTTCTGCAAA | ||
| + | ACGATTATGACAAACCGATGATTGAGAAACGCCTGGCCAAATTCACTGGTTTCGATGAATG | ||
| + | CCTGCTGTCTCAGTCTGGTTGGAATGCCAATGTTGGTCTGCTGCAAACAATCTGTCAGCCT | ||
| + | AACACGAACGTCTATATTGATTTCTTCGCCCACATGTCGCTGTGGGAAGGTGCTCGTTATG | ||
| + | CTAATGCTCAGGCCCATCCGTTTATGCACAACAACTGTGACCATCTGCGTATGCTGATTCA | ||
| + | GCGTCACGGTCCTGGTATTATCGTCGTGGACTCCATCTATTCTACCCTGGGGACCATTGCT | ||
| + | CCACTGGCTGAACTGGTGAATATCAGTAAAGAGTTTGGGTGTGCCCTGCTGGTTGATGAAA | ||
| + | GCCATTCTCTGGGAACCCATGGCCCGAACGGTGCCGGGCTGCTGGCGGAGCTGGGTCTGAC | ||
| + | ACGTGAAGTTCACTTCATGACCGCTTCGCTGGCAAAAACATTCGCCTATCGTGCTGGTGCC | ||
| + | ATTTGGTGTAACAACGAGGTTAATCGCTGTGTTCCGTTCATCTCTTATCCGGCCATCTTTA | ||
| + | GCAGTACACTGCTGCCGTATGAAGCTGCTGGTCTGGAAACAACCCTGGAGATTATCGAGTC | ||
| + | TGCCGATAACCGTCGTCAACATCTGGATCGTATGGCCCGTAAACTGCGTATTGGTCTGTCC | ||
| + | CAACTGGGTCTGACAATTCGTAGCGAATCTCAGATTATTGGCCTGGAGACTGGTGACGAGC | ||
| + | GTAATACCGAGAAAGTCCGTGATTATCTGGAGTCTAACGGCGTGTTTGGTAGCGTTTTTTG | ||
| + | TCGTCCGGCAACCTCTAAAAACAAAAACATCATCCGCCTGTCCCTGAATAGTGATGTGAAT | ||
| + | GATGAGCAGATCGCTAAAATCATTGAAGTCTGTTCGGATGCCGTGAATTATGGTGACTTCT | ||
| + | ATTTCCGCTGA </p> | ||
| - | |||
| - | + | <p>We then added the biobrick [http://partsregistry.org/Help:BioBrick_Prefix_and_Suffix prefix], promoter [http://partsregistry.org/Part:BBa_J23100 J23100], ribosome binding site [http://partsregistry.org/Part:BBa_B0034 B0034] before the CQSA sequence | |
| + | and the terminator [http://partsregistry.org/Part:BBa_B0015 B0015] and the biobrick [http://partsregistry.org/Help:BioBrick_Prefix_and_Suffix suffix], after the sequence. | ||
| + | </p> | ||
| - | + | We then sent these details of to Geneart and got them to do the hard work of synthesising our part for us. | |
| - | + | [[Image:CqsAinE.coli.png|600px]] | |
| - | + | <p>Once we received our DNA from synthesis we transformed it into E.coli and grew up some colonies. | |
| + | Unfortunately it is not currently possible to get parts synthesied and sent ready in the biobrick | ||
| + | so we made an over night broth culture of a selection of colonies and then miniprepped them the | ||
| + | following morning. We digested the resulting DNA with the restriction enzymes EcoR1 and Spe1 and | ||
| + | ran a gel to ensure that we had the part sizes that we wanted, we got further confirmation using | ||
| + | colony PCR with the [http://partsregistry.org/Part:BBa_G00100 VF2],[http://partsregistry.org/Part:BBa_G00101 VR] primer pair. </p> | ||
| - | + | We used gel extraction to get the parts of the size we wanted and ligated them into [http://partsregistry.org/Part:pSB1C3 pSB1C3]<br>we did more confirmation using digestion + gel and colony PCR and sent the samples which gave the nicest looking gel to the registry.<br> | |
| - | = | + | As you might imagine this is a very simplified account, for the a week by week account of our summer in the lab look at our [https://2010.igem.org/wiki/index.php?title=Team:St_Andrews/project/laboratory/notebook lab notebook.] |
| - | + | ||
| - | + | ||
| - | === | + | ===Reference=== |
| - | + | F Duan, J C March (2010) Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. PNAS 107-25 | |
Latest revision as of 01:15, 28 October 2010


Laboratory
Contents |
Introduction
The synthetic biology portion of our project has provided the undergraduate members of our team with a rare opportunity to work in the lab for an extended period of time. Many of us have gained important skills such as learning to use our time efficiently, planning the use of limited materials and organising experiments effectively.
Overview
This part of the lab work has focused on constructing two main biobricks - the bistable switch and the CAI-1 sender.
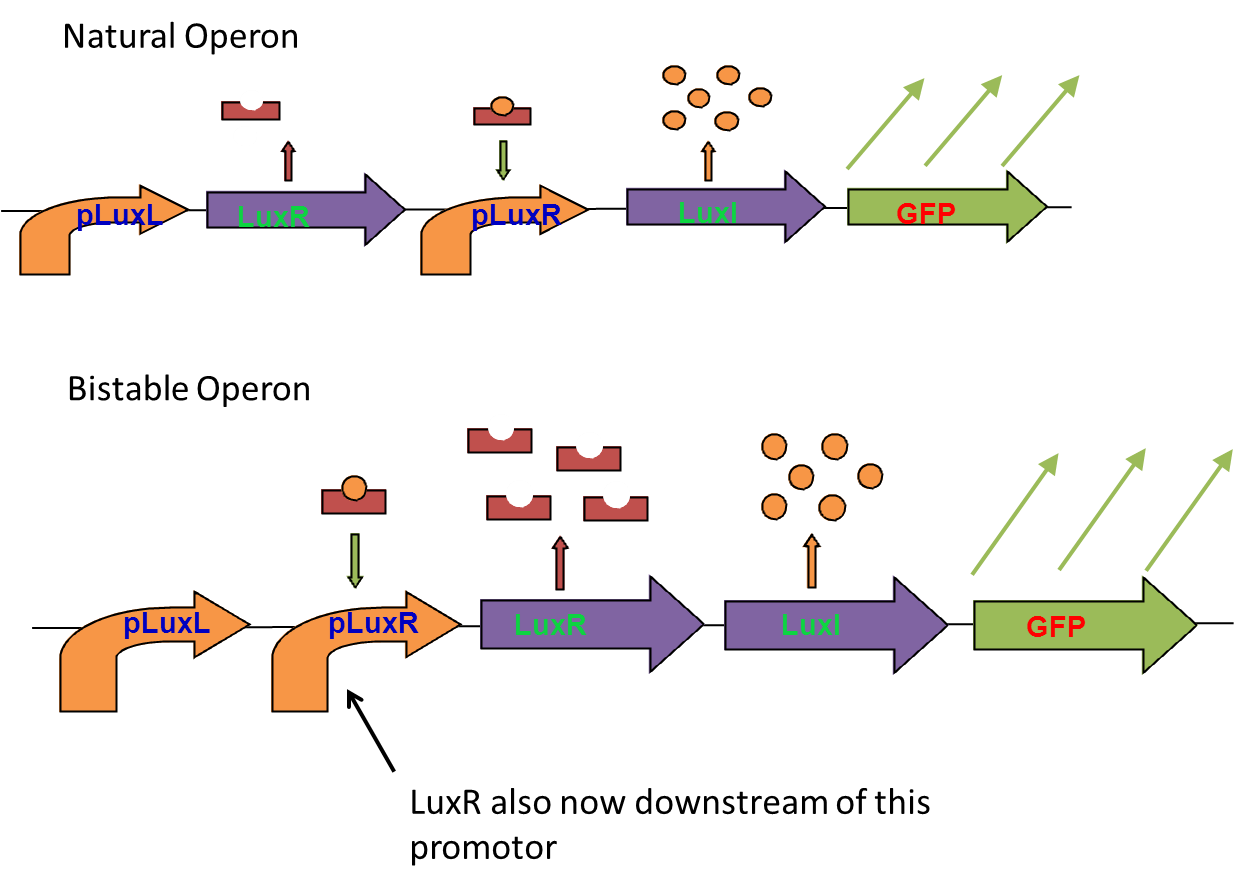
The bistable switch
To acheive bistable expression patterns in quorum sensing systems, we planned to rearrange the Lux operon so that both LuxI and LuxR were downstream of the Lux promotor.
To do this we planned to use already available parts from the registry to assemble our bistable operon.
We encountered problems with both 3 antibiotic assembly and In-Fusion PCR based assembly. These are detailed in the lab nootbook, but we eventually decided we had spent far too much time debugging our assembly methods and needed to move onto other areas of the project. The bistable operon was never finished.
Reference
EL Haseltine, FH Arnold (2007) - Implications of Rewiring Bacterial Quorum Sensing. Applied and Environmental Microbiology 74-2 437-445
The CAI-1 sender
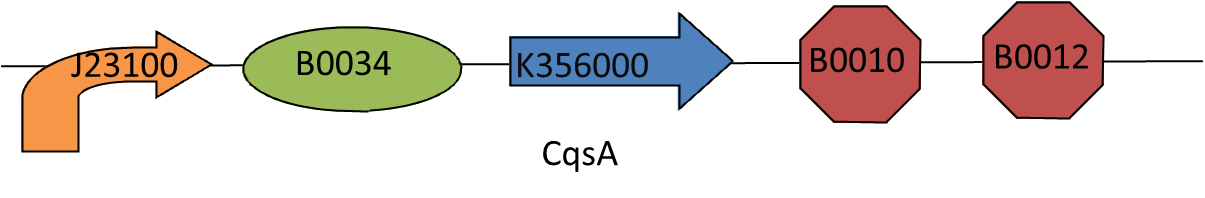
This biobrick is essential to our project. CAI-1, the cholera-specific auto inducing molecule, can switch off the production of cholera toxin by Vibrio cholerae when it is present in high concentrations. To make this biobrick the gene responsible for making the CAI-1 synthase (CqsA) was optimised for E. coli and then synthesised by geneart. CqsA can be found here.
First we found a sequence for the cholera gene for Vibrio cholerae autoinducer synthase.
the CAI-1 autoinducer synthase at genome.jp
We then codon optomised this gene for Escherichia coli using the functionality built into the Mr Gene Page
which gave us
ATGAACAAACCTCAGCTGCCTGACTTTATCCAAAACAAAATCGACCACTATATCGAGAACT ATTTCGACATTAACAAAAACGGCAAACACCTGGTGCTGGGCAAACAAGCATCACCGGATGA CATTATCCTGCAAAGCAACGACTATCTGGCCCTGGCTAATCACCCTCTGATCAAAGCTCGT CTGGCGAAAAGCCTGCTGGAAGAACAGCAATCCCTGTTTATGAGCGCCTCCTTTCTGCAAA ACGATTATGACAAACCGATGATTGAGAAACGCCTGGCCAAATTCACTGGTTTCGATGAATG CCTGCTGTCTCAGTCTGGTTGGAATGCCAATGTTGGTCTGCTGCAAACAATCTGTCAGCCT AACACGAACGTCTATATTGATTTCTTCGCCCACATGTCGCTGTGGGAAGGTGCTCGTTATG CTAATGCTCAGGCCCATCCGTTTATGCACAACAACTGTGACCATCTGCGTATGCTGATTCA GCGTCACGGTCCTGGTATTATCGTCGTGGACTCCATCTATTCTACCCTGGGGACCATTGCT CCACTGGCTGAACTGGTGAATATCAGTAAAGAGTTTGGGTGTGCCCTGCTGGTTGATGAAA GCCATTCTCTGGGAACCCATGGCCCGAACGGTGCCGGGCTGCTGGCGGAGCTGGGTCTGAC ACGTGAAGTTCACTTCATGACCGCTTCGCTGGCAAAAACATTCGCCTATCGTGCTGGTGCC ATTTGGTGTAACAACGAGGTTAATCGCTGTGTTCCGTTCATCTCTTATCCGGCCATCTTTA GCAGTACACTGCTGCCGTATGAAGCTGCTGGTCTGGAAACAACCCTGGAGATTATCGAGTC TGCCGATAACCGTCGTCAACATCTGGATCGTATGGCCCGTAAACTGCGTATTGGTCTGTCC CAACTGGGTCTGACAATTCGTAGCGAATCTCAGATTATTGGCCTGGAGACTGGTGACGAGC GTAATACCGAGAAAGTCCGTGATTATCTGGAGTCTAACGGCGTGTTTGGTAGCGTTTTTTG TCGTCCGGCAACCTCTAAAAACAAAAACATCATCCGCCTGTCCCTGAATAGTGATGTGAAT GATGAGCAGATCGCTAAAATCATTGAAGTCTGTTCGGATGCCGTGAATTATGGTGACTTCT ATTTCCGCTGA
We then added the biobrick prefix, promoter J23100, ribosome binding site B0034 before the CQSA sequence and the terminator B0015 and the biobrick suffix, after the sequence.
We then sent these details of to Geneart and got them to do the hard work of synthesising our part for us.
Once we received our DNA from synthesis we transformed it into E.coli and grew up some colonies. Unfortunately it is not currently possible to get parts synthesied and sent ready in the biobrick so we made an over night broth culture of a selection of colonies and then miniprepped them the following morning. We digested the resulting DNA with the restriction enzymes EcoR1 and Spe1 and ran a gel to ensure that we had the part sizes that we wanted, we got further confirmation using colony PCR with the VF2,VR primer pair.
We used gel extraction to get the parts of the size we wanted and ligated them into pSB1C3
we did more confirmation using digestion + gel and colony PCR and sent the samples which gave the nicest looking gel to the registry.
As you might imagine this is a very simplified account, for the a week by week account of our summer in the lab look at our lab notebook.
Reference
F Duan, J C March (2010) Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. PNAS 107-25
 "
"