Team:SDU-Denmark/project-p
From 2010.igem.org
(Difference between revisions)
(→K343004) |
|||
| Line 10: | Line 10: | ||
----- | ----- | ||
==== K343007 ==== | ==== K343007 ==== | ||
| + | <p style="text-align: justify;"> | ||
| + | <br> | ||
The part K343003 (from now on shortly called PS) is a generator for the SopII-HtrII photosensor from Natronomonas Pharaonis coupled to E.Colis chemotaxis pathway via the Salmonella protein Tar. This part's effect on the system is to make E.Coli phototactic, so that it becomes aware of different light conditions. So for characterization of this part we made examination of motility and motility patterns our first priority and plasmid stability and growth of the cells our second. <br> | The part K343003 (from now on shortly called PS) is a generator for the SopII-HtrII photosensor from Natronomonas Pharaonis coupled to E.Colis chemotaxis pathway via the Salmonella protein Tar. This part's effect on the system is to make E.Coli phototactic, so that it becomes aware of different light conditions. So for characterization of this part we made examination of motility and motility patterns our first priority and plasmid stability and growth of the cells our second. <br> | ||
There is a wide range of motility assays for chemotaxis in bacteria, which meant that we had a broad spectrum of experiments to choose from, which just had to be tweaked for making them suited for the analysis of phototaxis. The two experiments we chose for analysing the effect of this part (PS), were growth of the bacterial cultures in semi-solid agar and computer analysis of swimming motility through video microscopy:<br> | There is a wide range of motility assays for chemotaxis in bacteria, which meant that we had a broad spectrum of experiments to choose from, which just had to be tweaked for making them suited for the analysis of phototaxis. The two experiments we chose for analysing the effect of this part (PS), were growth of the bacterial cultures in semi-solid agar and computer analysis of swimming motility through video microscopy:<br> | ||
| Line 39: | Line 41: | ||
When played at real time speed these videos seem slow and not showing any tendency, though when played at 4 times the normal speed, the photosensor will seem to show slightly increased motility when exposed to blue light. Even though the bacteria displayed disappointing motility, we went on and did computer analysis on the videos by the help of the open source software "CellTrack" (Link!). The paths that were mapped showed a longer distance traveling for the photosensor exposed to blue light, when compared with the rest of the samples. This could indicated a decreased tumbling frequency, but since the bacteria's motility was very low, the results were stamped as unreliable and a new protocol had to be devised.<br><br> | When played at real time speed these videos seem slow and not showing any tendency, though when played at 4 times the normal speed, the photosensor will seem to show slightly increased motility when exposed to blue light. Even though the bacteria displayed disappointing motility, we went on and did computer analysis on the videos by the help of the open source software "CellTrack" (Link!). The paths that were mapped showed a longer distance traveling for the photosensor exposed to blue light, when compared with the rest of the samples. This could indicated a decreased tumbling frequency, but since the bacteria's motility was very low, the results were stamped as unreliable and a new protocol had to be devised.<br><br> | ||
'''Computerized analysis of the bacterial motility with the Thor prototype by Unisensor A/S and the Unify software:''' | '''Computerized analysis of the bacterial motility with the Thor prototype by Unisensor A/S and the Unify software:''' | ||
| + | <br> | ||
| + | </p> | ||
==== K343004 ==== | ==== K343004 ==== | ||
| Line 45: | Line 49: | ||
The FlhDC operon is the master regulator of flagella synthesis. a more detailed description of the operon can be found [https://2010.igem.org/Team:SDU-Denmark/project-t#Hyperflagellation here] <br> | The FlhDC operon is the master regulator of flagella synthesis. a more detailed description of the operon can be found [https://2010.igem.org/Team:SDU-Denmark/project-t#Hyperflagellation here] <br> | ||
The purpose of this composite part in our system is to hyper flagellate our cells so that a grater force can be created in the microtubes. The FlhDC operon is naturally found in the ''E. coli'' strain MG1655 genome. We extracted the operon and incearted a silent mutation (T to C) at position 822 in the operon because without the mutation this site is a Pst1 site, and it would therefor constitute problems when assembling the composit part. <br> We have made three FlhDC parts:<br> [http://partsregistry.org/Part:BBa_K343100 K343100] is the coding sequence of the native FlhDC operon with the Pst1 site <br> [http://partsregistry.org/Part:BBa_K343000 K343000] is the coding sequence of the mutated FlhDC operon <br> [http://partsregistry.org/Part:BBa_K343004 K343004] is the composite part containing a TetR repressable promoter (constitutive when no TetR is pressent)+ an RBS (J13002), the K343000 part and a double terminator (B0015). <br> | The purpose of this composite part in our system is to hyper flagellate our cells so that a grater force can be created in the microtubes. The FlhDC operon is naturally found in the ''E. coli'' strain MG1655 genome. We extracted the operon and incearted a silent mutation (T to C) at position 822 in the operon because without the mutation this site is a Pst1 site, and it would therefor constitute problems when assembling the composit part. <br> We have made three FlhDC parts:<br> [http://partsregistry.org/Part:BBa_K343100 K343100] is the coding sequence of the native FlhDC operon with the Pst1 site <br> [http://partsregistry.org/Part:BBa_K343000 K343000] is the coding sequence of the mutated FlhDC operon <br> [http://partsregistry.org/Part:BBa_K343004 K343004] is the composite part containing a TetR repressable promoter (constitutive when no TetR is pressent)+ an RBS (J13002), the K343000 part and a double terminator (B0015). <br> | ||
| - | + | The composite part is caracterized firstly by using a motility assay and secondly by measuring plasmid stability and cell growth. <br><br> | |
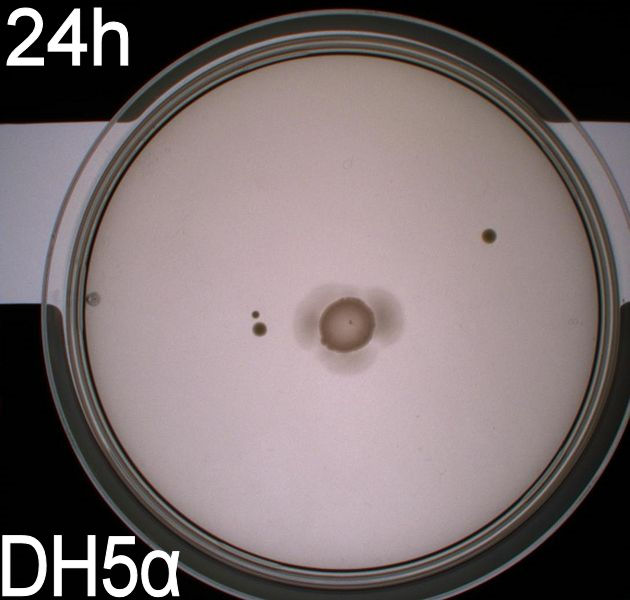
| - | '''1. Growth of bacterial culture on semi-solid agar plates | + | '''1. Motility assay (Experiment 1 with WT and negative control):''' <br> Growth of bacterial culture on semi-solid agar plates <br> |
[[Image:Team-SDU-Denmark-Flagellamotility-exp1-DH5a.JPG|250px|DH5a]] | [[Image:Team-SDU-Denmark-Flagellamotility-exp1-DH5a.JPG|250px|DH5a]] | ||
[[Image:Team-SDU-Denmark-Flagellamotility-exp1-MG1655.JPG|250px|MG1655]]<br><br> | [[Image:Team-SDU-Denmark-Flagellamotility-exp1-MG1655.JPG|250px|MG1655]]<br><br> | ||
| Line 53: | Line 57: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
| + | ''' 2. Motility assay (doublicate with WT, negative- and positive-control):''' <br> Growth of bacterial culture on semi-solid agar plates <br> | ||
| + | ''' 3. Stability assay '''<br> | ||
| + | ''' 4. Growth measurment ''' <br> | ||
<br><br> | <br><br> | ||
Revision as of 11:53, 24 October 2010
 "
"