Team:Newcastle/Non-target-environment kill switch
From 2010.igem.org
PhilipHall (Talk | contribs) |
PhilipHall (Talk | contribs) (→Non-target environment detection) |
||
| (3 intermediate revisions not shown) | |||
| Line 15: | Line 15: | ||
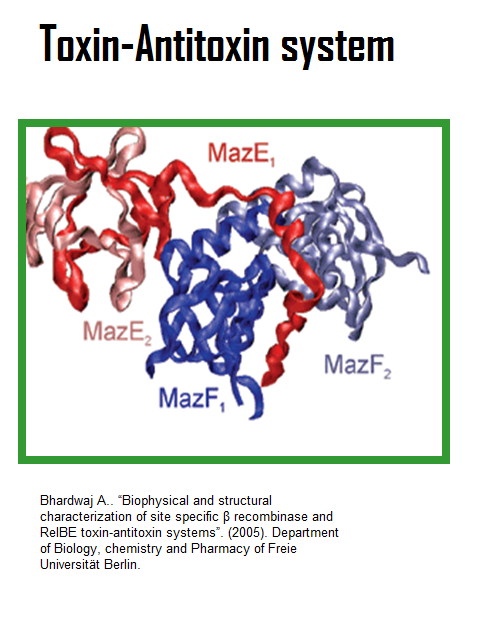
=''mazEF''= | =''mazEF''= | ||
| - | We have designed the a kill switch which uses the | + | We have designed the a kill switch which uses the ''mazEF'' Toxin-Antitoxin system: |
[[Image:TeamNewcastleMazEF.png]] | [[Image:TeamNewcastleMazEF.png]] | ||
| - | + | ''mazEF'' is found in strains of ''Escherichia coli'' and ''Bacillus subtilis''. | |
Consists of two two adjacent genes, ''mazE'' and ''mazF'', located downstream from the ''relA'' gene. | Consists of two two adjacent genes, ''mazE'' and ''mazF'', located downstream from the ''relA'' gene. | ||
| - | ''mazF'' encodes a stable toxin, | + | ''mazF'' encodes a stable toxin, ''mazF''. |
| - | ''mazE'' encodes a labile antitoxin, | + | ''mazE'' encodes a labile antitoxin, ''mazE'', degraded in vivo by the ATP-dependent ClpPA serine protease. |
| - | + | ''mazE'' and ''mazF'' are coexpressed and mazEF is negatively auto-regulated at the level of transcription by the combined action of both ''mazE'' and ''mazF'' proteins on the ''mazEF'' promoter P2. | |
| - | This system is activated by several stressful conditions that prevent the expression of mazEF, and thereby | + | This system is activated by several stressful conditions that prevent the expression of ''mazEF'', and thereby ''mazE'' synthesis, and thus trigger cell death. These conditions include: |
* extreme amino acid starvation leading to the production of the starvation-signaling molecule ppGpp | * extreme amino acid starvation leading to the production of the starvation-signaling molecule ppGpp | ||
* inhibition of transcription and/or translation by antibiotics including rifampicin, chloramphenicol, and spectinomycin | * inhibition of transcription and/or translation by antibiotics including rifampicin, chloramphenicol, and spectinomycin | ||
* Doc protein, a general inhibitor of translation which is the toxic product of the ‘‘addiction module’’ phd-doc of the plasmid prophage P1. | * Doc protein, a general inhibitor of translation which is the toxic product of the ‘‘addiction module’’ phd-doc of the plasmid prophage P1. | ||
| + | |||
[http://partsregistry.org/Cell_death| More cell death parts on the parts registry ] | [http://partsregistry.org/Cell_death| More cell death parts on the parts registry ] | ||
| Line 36: | Line 37: | ||
=Non-target environment detection= | =Non-target environment detection= | ||
| - | In our [http://partsregistry.org/Part:BBa_K302035 | + | In our [http://partsregistry.org/Part:BBa_K302035| construct] a sucrose inducer is placed in the construct so that ''mazE'' and ''mazF'' can be switched off by a depletion of of sucrose in the media, be this running out of it after achieving its purpose in the microcrack, or if it is sprayed into the environment, where there is little or no sucrose (e.g in the soil, on human skin). The deactivation of expression will lead to a build up of ''mazF'' and the death of the cell |
| + | |||
| + | =References= | ||
| + | |||
| + | Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. (2006) Bacterial programmed cell death and multicellular behavior in bacteria. ''PLoS Genet.'' 2: e135 | ||
| + | |||
| + | Pellegrini, O., Mathy, N., Gogos, A., Shapiro, L., and Condon, C. (2005) The ''Bacillus subtilis'' ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. ''Mol Microbiol'' 56: 1139–1148. | ||
| + | |||
| + | Shimotsu, H., and Henner, D. J. (1986) ''J. Bacteriol.'' 168, 380–388 | ||
| - | |||
| - | |||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Latest revision as of 02:08, 26 October 2010

| |||||||||||||
| |||||||||||||
Contents |
Kill switch
We have considered a kill switch which in the absence of key nutrient would activate and help stop the spread of our bacteria.
Ideas
- A kill switch could be designed to take advantage of the subtilin lantibiotic we are using for cell-cell communication by switching off the immunity genes.
- pH (and other differences in conditions) could be factored into the control of our kill switch, for example due to a pH sensitive promoter.
Holin-Lysozyme lysis
UC Berkeley developed a Holin-Lysozyme lysis device in 2008.
mazEF
We have designed the a kill switch which uses the mazEF Toxin-Antitoxin system:
mazEF is found in strains of Escherichia coli and Bacillus subtilis. Consists of two two adjacent genes, mazE and mazF, located downstream from the relA gene. mazF encodes a stable toxin, mazF.
mazE encodes a labile antitoxin, mazE, degraded in vivo by the ATP-dependent ClpPA serine protease.
mazE and mazF are coexpressed and mazEF is negatively auto-regulated at the level of transcription by the combined action of both mazE and mazF proteins on the mazEF promoter P2.
This system is activated by several stressful conditions that prevent the expression of mazEF, and thereby mazE synthesis, and thus trigger cell death. These conditions include:
- extreme amino acid starvation leading to the production of the starvation-signaling molecule ppGpp
- inhibition of transcription and/or translation by antibiotics including rifampicin, chloramphenicol, and spectinomycin
- Doc protein, a general inhibitor of translation which is the toxic product of the ‘‘addiction module’’ phd-doc of the plasmid prophage P1.
[http://partsregistry.org/Cell_death| More cell death parts on the parts registry ]
Non-target environment detection
In our [http://partsregistry.org/Part:BBa_K302035| construct] a sucrose inducer is placed in the construct so that mazE and mazF can be switched off by a depletion of of sucrose in the media, be this running out of it after achieving its purpose in the microcrack, or if it is sprayed into the environment, where there is little or no sucrose (e.g in the soil, on human skin). The deactivation of expression will lead to a build up of mazF and the death of the cell
References
Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. (2006) Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2: e135
Pellegrini, O., Mathy, N., Gogos, A., Shapiro, L., and Condon, C. (2005) The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol Microbiol 56: 1139–1148.
Shimotsu, H., and Henner, D. J. (1986) J. Bacteriol. 168, 380–388
 
|
 "
"