Team:Newcastle/Initial filamentous
From 2010.igem.org
RachelBoyd (Talk | contribs) (→Filamentous Cells) |
Swoodhouse (Talk | contribs) |
||
| (3 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| - | |||
| - | =Filamentous Cells= | + | =Initial research: Filamentous Cells= |
| + | |||
| + | We have chosen to produce a filamentous cell phenotype to strengthen the cracks found in concrete. Our initial research lead us to look at the genes and systems listed below. We eventually decided on overexpressing the ''yneA'' because we think it will produce the best phenotype and only requires we control one gene. Another advantage is its small gene size. | ||
| + | |||
| + | ''Bacillus subtilis'' in response to stress such as DNA damage stops the cells from dividing. This is a part of the SOS response initiated by the accumulation of single stranded DNA from DNA damage or stalled replication. Two proteins are vital for this response: RecA and LexA. RecA forms filaments on ssDNA and promotes the autocleavage of LexA. LexA usually represses the SOS operon. ''dinR'' is homologous to ''lexA'' in ''E. coli'' and is transcribed in the opposite direction of ''yneA''. | ||
| + | |||
| + | SOS response is believe to be a universal bacteria phenomenon first studied in ''E.coli'' -''lexA'', ''recA'' | ||
| + | |||
| + | In ''Bacillus subtillis'' (gram positive) ''dinR'' protein is homologous to ''lexA'' (Repressor of ''din''-damage inducible genes). ''din'' genes include ''uvrA'', ''uvrB'', ''dinB'', ''dinC'' ''dinR'' and ''recA''. DNA damage inhibits cell division. | ||
| + | |||
| + | {| | ||
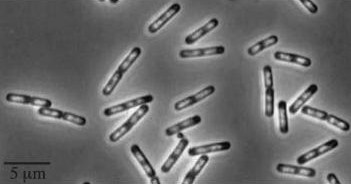
| + | !Wild type ''Bacillus subtilis'' | ||
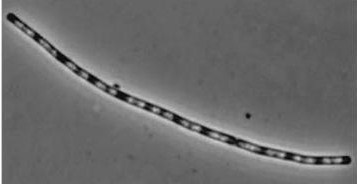
| + | !''dinR''KO Mutant | ||
| + | |- | ||
| + | ![[Image:Wild type Bacillus subtillis.jpg]] | ||
| + | ![[Image:dinR KO.jpg]] | ||
| + | |} | ||
| + | |||
| + | '''Figure1''': The images above show ''Bacillus subtilis'' Wild type and ''dinR''KO mutant, and the change in cell length. ''dinR'' KO mutant over expresses the divergent (opposite direction) transcript for YneA, YneB and YnzC. These genes form the SOS regulon (''recA'' independent SOS response). | ||
| + | |||
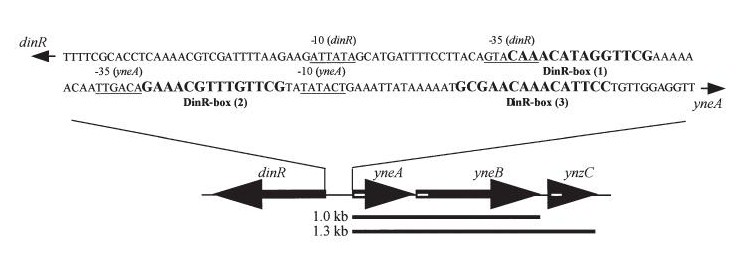
| + | [[Image:Coding region.jpg]] | ||
| + | |||
| + | |||
| + | '''Figure2''':The diagram above shows the Coding region for ''dinR'' and ''yneA'' showing divergent expression. | ||
| + | |||
| + | Expression of YneA from IPTG controlled promoter in wildtype leads to elongation. | ||
| + | Disruption of YneA in SOS response leads to reduced elongation. Altering YneB and YnzC expression does not affect cell morphology. | ||
| + | |||
| + | {| | ||
| + | |- | ||
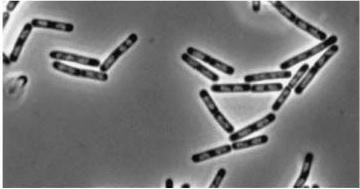
| + | |[[Image:Double Mutant.jpg|Double mutant (''dinR'' YneA)]] | ||
| + | |[[Image:yneA_ftsZ.png|!Graph showing ''yneA'' expression correlated with FtsZ ring formation and cells length]] | ||
| + | |} | ||
| + | |||
| + | '''Figure3'''(above left): Shows the double mutant ''dinR'' overexpression cancels out the filament formation via over expression of ''yneA''. | ||
| + | |||
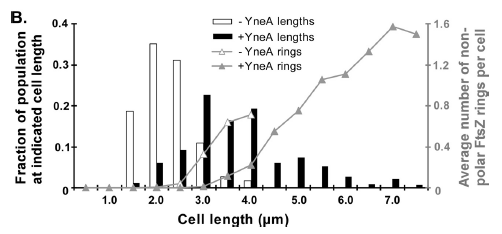
| + | '''Figure4'''(above right):This graph shows the correllation between reduced FtsZ ring formation, increased cell length and overexpression of ''yneA''. | ||
| + | |||
| + | YneA protein required to suppress cell division and not chromosome replication or segregation. | ||
| + | |||
| + | FtsZ is important for bacterial cell division forming a ring structure at the division site by polymerising | ||
| + | assembling other proteins necessary for division at the site. | ||
| + | |||
| + | FtsZ localises to the cell division cycle unless ''dinR'' is disrupted or YneA is being induced. | ||
| + | YneA suppresses FtsZ ring formation which is proven by 2 hybrid protein association test. | ||
| + | |||
| + | YneA expression by the inactivation of ''dinR'' by RecA is important. | ||
| + | |||
| + | |||
==Filamentous cells genes list== | ==Filamentous cells genes list== | ||
| Line 16: | Line 63: | ||
*Inhibitors of Daughter cell separation: ''lytC,D,E,F and cwlS'' *Chains rather than filaments, ''yneA'' is also reported to increase the time spent in chains well into the stationary phase of bacterial growth. | *Inhibitors of Daughter cell separation: ''lytC,D,E,F and cwlS'' *Chains rather than filaments, ''yneA'' is also reported to increase the time spent in chains well into the stationary phase of bacterial growth. | ||
| - | == | + | |
| + | ==Alternative system: MinCDJ== | ||
{| | {| | ||
Latest revision as of 16:28, 26 October 2010

| |||||||||||||
| |||||||||||||
Initial research: Filamentous Cells
We have chosen to produce a filamentous cell phenotype to strengthen the cracks found in concrete. Our initial research lead us to look at the genes and systems listed below. We eventually decided on overexpressing the yneA because we think it will produce the best phenotype and only requires we control one gene. Another advantage is its small gene size.
Bacillus subtilis in response to stress such as DNA damage stops the cells from dividing. This is a part of the SOS response initiated by the accumulation of single stranded DNA from DNA damage or stalled replication. Two proteins are vital for this response: RecA and LexA. RecA forms filaments on ssDNA and promotes the autocleavage of LexA. LexA usually represses the SOS operon. dinR is homologous to lexA in E. coli and is transcribed in the opposite direction of yneA.
SOS response is believe to be a universal bacteria phenomenon first studied in E.coli -lexA, recA
In Bacillus subtillis (gram positive) dinR protein is homologous to lexA (Repressor of din-damage inducible genes). din genes include uvrA, uvrB, dinB, dinC dinR and recA. DNA damage inhibits cell division.
| Wild type Bacillus subtilis | dinRKO Mutant |
|---|---|
| 
|
Figure1: The images above show Bacillus subtilis Wild type and dinRKO mutant, and the change in cell length. dinR KO mutant over expresses the divergent (opposite direction) transcript for YneA, YneB and YnzC. These genes form the SOS regulon (recA independent SOS response).
Figure2:The diagram above shows the Coding region for dinR and yneA showing divergent expression.
Expression of YneA from IPTG controlled promoter in wildtype leads to elongation. Disruption of YneA in SOS response leads to reduced elongation. Altering YneB and YnzC expression does not affect cell morphology.
| 
|
Figure3(above left): Shows the double mutant dinR overexpression cancels out the filament formation via over expression of yneA.
Figure4(above right):This graph shows the correllation between reduced FtsZ ring formation, increased cell length and overexpression of yneA.
YneA protein required to suppress cell division and not chromosome replication or segregation.
FtsZ is important for bacterial cell division forming a ring structure at the division site by polymerising assembling other proteins necessary for division at the site.
FtsZ localises to the cell division cycle unless dinR is disrupted or YneA is being induced. YneA suppresses FtsZ ring formation which is proven by 2 hybrid protein association test.
YneA expression by the inactivation of dinR by RecA is important.
Filamentous cells genes list
- yneA (Transcribed with yneB, ynzC, is an analogue of sulA in E.coli) * our biobrick is designed to over express this gene reducing cell division possibly by inhibiting FtsZ ring formation or constriction.
- dinR (Homologue of lexA in E.coli transcribed in the opposite direction)
- ftsZ (Involved in the recruitment of other proteins to the divisisome for cytokinesis, strangely over expression results in disruption of Zring formation as well as reduced expression)
- secA (Involved in the secretion of extracellular proteins and the insertion of transmembrane proteins)
- recA (Involved in SOS response removing the repressor DinR (LexA))
- wpr and epr produce extracellular proteases that cleave the signal peptide/transmembrane domain of YneA
- ezr produces a protein which sequesters FtsZ monomer by binding its C terminal domain and also inhibits GTP binding; however overexpression does not result in filamentation.
- min C,D ,J and divIVA prevent polar cell division .
- Positive regulators of FtsZ: ftsA, zapA, zipA, ftsL and divIC
- Inhibitors of Daughter cell separation: lytC,D,E,F and cwlS *Chains rather than filaments, yneA is also reported to increase the time spent in chains well into the stationary phase of bacterial growth.
Alternative system: MinCDJ
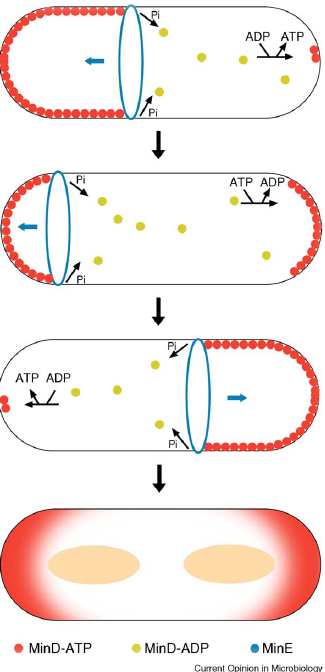
| In B.subtilis cell division occurs precisely mid cell via the formation of the FtsZ ring (tubulin homologue), through Noc (Nucleoid occlusion: prevents division over nucleoids) and the min system (well described in E.coli), which prevents division taking place at the ends of the cell (poles). |
| Main function of Min system to prevent mini cell formation and ensuring only one cell division occurs per cell cycle. |
| Min’s role is not just in the inhibition of FtsZ. FtsZ recruits other components causing synthesis of the new wall and cell invagination. |
|
| Cell division is regulated spatially and temporally. |
| Min C inhibits FtsZ ring formation; Min C interacts with Min D via its C-terminal domain. Min C inhibits lateral interaction between the filaments. |
| Min D is a membrane associated ATPase. (Min E ensures high concentrations of MinCD at the poles in E.coli, no Min E homologue in B.subtilis! Instead DivIVA acts as a topological factor). |
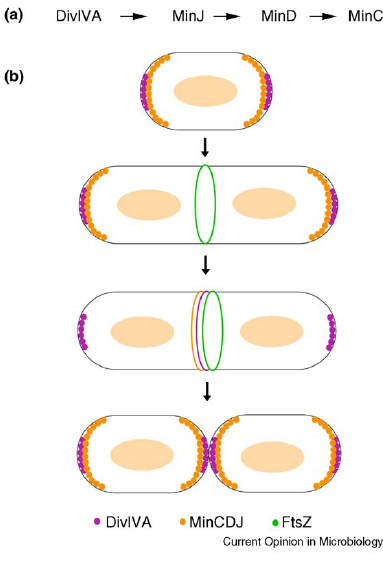
| Min C in B.subtilis is responsible for localisation (shown using GFP) showed a primary site of localisation at the site of active division challenging the original model for the role of Min. |
| Min J was discovered linking Min D to DivIVA and therefore necessary for localisation. |
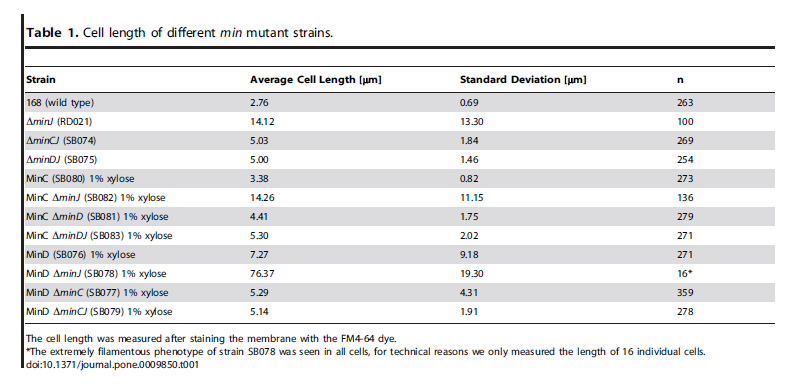
| Over expression of Min D in the absence of Min J causes lethal filamentation. |
|
| Min C sequence |
| http://www.ncbi.nlm.nih.gov/nuccore/NC_000964.3?from=2858550&to=2859298&strand=true&report=graph&content=5 |
| http://www.ncbi.nlm.nih.gov/gene/937500 |
| MinD sequence |
| http://www.ncbi.nlm.nih.gov/nuccore/NC_000964.3?from=2857736&to=2858622&strand=true&report=graph&content=5 |
| http://www.ncbi.nlm.nih.gov/gene/937499 |
| MinJ sequence |
| http://www.ncbi.nlm.nih.gov/nuccore/NC_000964.3?from=3620287&to=3621598&strand=true&report=graph&content=5 |
| http://www.ncbi.nlm.nih.gov/gene/936668 |
| van Baarle, S., & Bramkamp, M. (2010). The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly. PloS one, 5(3), e9850. doi: 10.1371/journal.pone.0009850. |
| Bramkamp, M. & van Baarle, S., 2009. Division site selection in rod-shaped bacteria. Current opinion in microbiology, 12(6), 683-8. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19884039. |
 "
"