Team:Imperial College London/Results
From 2010.igem.org
(→Cat-O2-lase Protein) |
|||
| Line 7: | Line 7: | ||
=Cat-O2-lase Protein= | =Cat-O2-lase Protein= | ||
| - | The XylE gene, originating from the TOL plasmid of Pseudomonas putida, was the choice for the output signal mediator of our system. The system is based on | + | The XylE gene, originating from the TOL plasmid of Pseudomonas putida, was the choice for the output signal mediator of our system. The system is based on catechol 2,3-dioxygenase enzyme. Colonies of cells that express xylE gene product become yellow within seconds after selection plates are sprayed with catechol, a colorless substrate, that is converted by CatO2ase to the yellow product, 2-hydroxymuconic semialdehyde. This reporter system is ideal for our project as it has very fast kinetics, gives a visual output by naked eye and the substrate used is very cheap. |
==Characterization of CatO2lase protein== | ==Characterization of CatO2lase protein== | ||
Revision as of 21:09, 20 October 2010
| Results |
Cat-O2-lase Protein
The XylE gene, originating from the TOL plasmid of Pseudomonas putida, was the choice for the output signal mediator of our system. The system is based on catechol 2,3-dioxygenase enzyme. Colonies of cells that express xylE gene product become yellow within seconds after selection plates are sprayed with catechol, a colorless substrate, that is converted by CatO2ase to the yellow product, 2-hydroxymuconic semialdehyde. This reporter system is ideal for our project as it has very fast kinetics, gives a visual output by naked eye and the substrate used is very cheap.
Characterization of CatO2lase protein
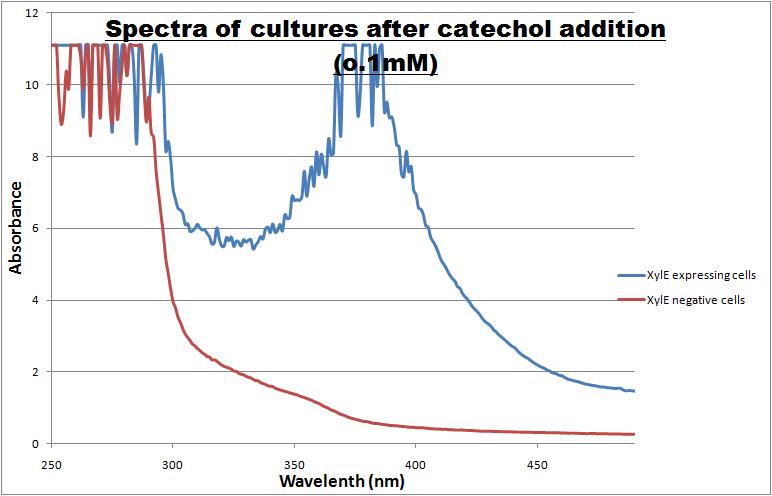
The CatO2lase protein, except as an ideal output signal for our engineered bacterial detector it can also serve as a great reporter gene. Its characteristics can be exploited in a wide range of fields across biological sciences field, so it was one of the team's first candidates for further characterization. Qualitative and quantitative data were gathered and presented. All further tests involving XylE transformed cells were quantified by measuring absorbonce at 380nm wavelenth.
Catechol ,the initial substrate of CatO2las enzyme, is colourless. However within seconds of its addition the colonies/liquid cultures of XylE expressing cells become yellow, indicating production of product which absorbs light in the region visible spectrum. An spectrophotometric assay was prepared, where the spectra of two cultures of E.coli (one XylE gene transformed and the other not)were compared on addition of 0.1mM Catechol substrate. The spectra (figure 1) showed that in XylE transformed cells, a broad peak appears at about 380nm. The absorbance of the particular wavelengths of light by the product of the enzymatic reaction 2-hydroxymuconic semialdehyde, is what causes the yellow output.
References
- Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Zukowski, M.M., Gaffney, D.F., Speck, D., Kauffmann, M., Findeli, A., Wisecup, A., Lecocq, J.P. Proc. Natl. Acad. Sci. U.S.A. (1983)
 "
"