Team:Freiburg Bioware/NoteBook/Labjournal/October2
From 2010.igem.org
(→Cloning of lITR_CMV_betaglobin and lITR_phTERT_betaglobin into pSB1C3_CD) |
(→147. labday 12.10.2010) |
||
| Line 215: | Line 215: | ||
Perfomance of another try by harvesting viruses of the cell pellet with RIPA buffer & freeze/ thaw. <br/> | Perfomance of another try by harvesting viruses of the cell pellet with RIPA buffer & freeze/ thaw. <br/> | ||
Western Blot of unmodified, and modified capsids (N-terminal fusion and VP1 insertion with CFP ref. mVenus) --> size shift should be detectable.<br/> | Western Blot of unmodified, and modified capsids (N-terminal fusion and VP1 insertion with CFP ref. mVenus) --> size shift should be detectable.<br/> | ||
| + | |||
| + | |||
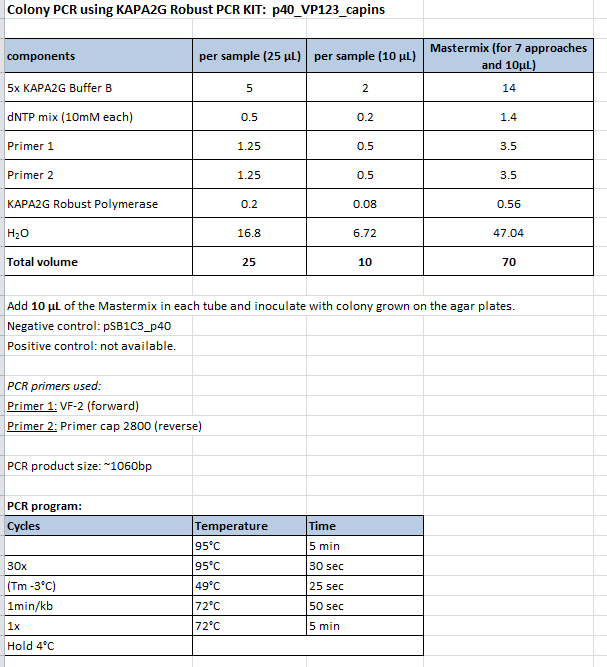
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Mini-Prep and test digestion of several constructs</b></p>==== | ||
| + | <b>Investigator: Stefan</b><br /> | ||
| + | |||
| + | Glycerol stocks were prepared:<br> | ||
| + | |||
| + | <ul> | ||
| + | <li>B651 = pSB1C3_P40_VP123</li> | ||
| + | <li>B652 = pSB1C3_litr_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR</li> | ||
| + | <li>B653 = pSB1C3_litr_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR</li> | ||
| + | <li>B654 = pSB1C3_Iitr_CMV_ß-globin_mGMK_TK30_hgH_rITR</li> | ||
| + | <li>B655 = pSB1C3_Iitr_CMV_ß-globin_mGMK_TK30_hgH_rITR</li> | ||
| + | <li>B656 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO</li> | ||
| + | <li>B657 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO</li> | ||
| + | <li>B658 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO</li> | ||
| + | <li>B659 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO</li> | ||
| + | <li>B660 = pSB1C3_litr_phTERT_ß-globin_cfp_hgH-ritr</li> | ||
| + | <li>B661 = pSB1C3_litr_phTERT_ß-globin_cfp_hgH-ritr</li> | ||
| + | </ul><br> | ||
| + | |||
| + | Mini-Prep was performed according to standard protocol:<br> | ||
| + | |||
| + | <ul> | ||
| + | <li>P821 = pSB1C3_P40_VP123 c = 327,4 ng/µl</li> | ||
| + | <li>P822 = pSB1C3_litr_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR c = 342,9 ng/µl</li> | ||
| + | <li>P823 = pSB1C3_litr_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR c = 324,0 ng/µl</li> | ||
| + | <li>P824 = pSB1C3_Iitr_CMV_ß-globin_mGMK_TK30_hgH_rITR c = 81,6 ng/µl</li> | ||
| + | <li>P825 = pSB1C3_Iitr_CMV_ß-globin_mGMK_TK30_hgH_rITR c = 148,4 ng/µl</li> | ||
| + | <li>P826 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO c = 105,8 ng/µl</li> | ||
| + | <li>P827 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO c = 121,1 ng/µl</li> | ||
| + | <li>P828 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO c = 188,4 ng/µl</li> | ||
| + | <li>P829 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO c = 140,4 ng/µl</li> | ||
| + | <li>P830 = pSB1C3_litr_phTERT_ß-globin_cfp_hgH-ritr c = 197,2 ng/µl</li> | ||
| + | <li>P831 = pSB1C3_litr_phTERT_ß-globin_cfp_hgH-ritr c = 207,3 ng/µl</li> | ||
| + | </ul> | ||
| + | |||
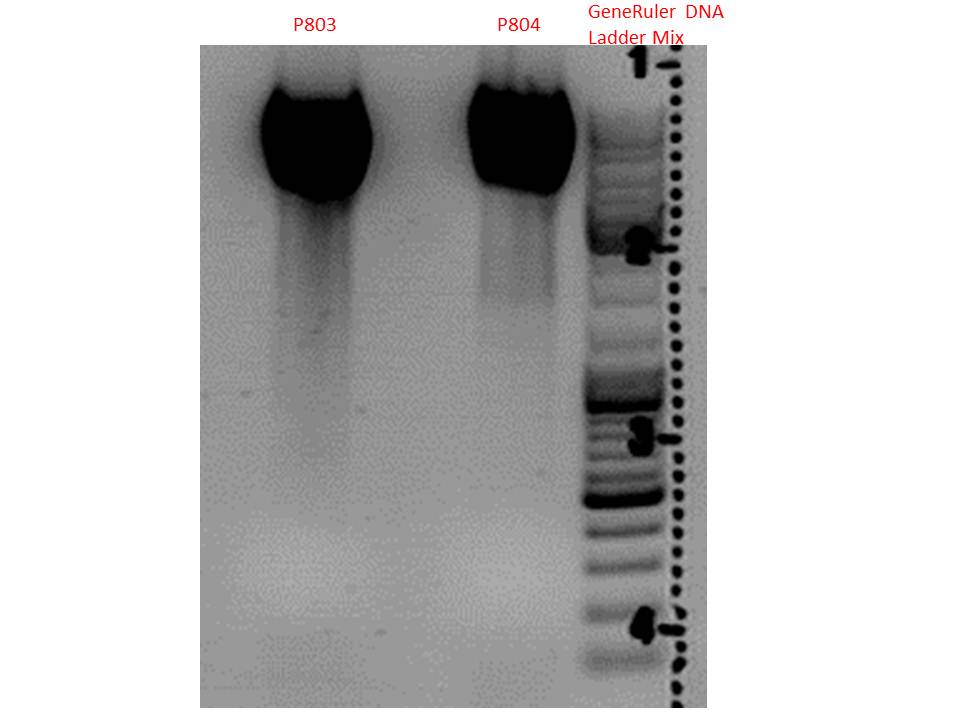
| + | Test digestion:<br> | ||
| + | {| border="1" | ||
| + | | align="left" | '''Components''' ||align="left"| <b>P803 + P804 / µl</b> | ||
| + | |- | ||
| + | | align="left" | DNA ||align="left"| 2,5 | ||
| + | |- | ||
| + | | align="left" | Buffer 4 ||align="left"| 1 | ||
| + | |- | ||
| + | | align="left" | BSA (10x) ||align="left"| 1 | ||
| + | |- | ||
| + | | align="left" | PstI ||align="left"| 0,3 | ||
| + | |- | ||
| + | | align="left" | H<sub>2</sub>O ||align="left"| 5,2 | ||
| + | |- | ||
| + | | align="left" | '''Total volume''' ||align="left"| <b>10</b> | ||
| + | |} | ||
| + | |||
| + | Gel:<br> | ||
| + | 0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes<br> | ||
| + | |||
| + | [[Image:Freiburg10 test digestion 101010.jpg|550px]] | ||
| + | <p style="color:#66bbff;"><i>Comment</i>: .</p> | ||
===<p style="font-size:17px; background-color:#00dd77;">148. labday 13.10.2010</p>=== | ===<p style="font-size:17px; background-color:#00dd77;">148. labday 13.10.2010</p>=== | ||
Revision as of 15:48, 12 October 2010
- March (labday 1)
- April (labday 2 - 5)
- May (labday 6 - 17)
- June (labday 18 - 45)
- July (labday 46 - 75)
- August part 1 (labday 76 - 92)
- August part 2 (labday 93 - 106)
- September part 1 (labday 107 - 123)
- September part 2 (labday 124 - 135)
- October part 1 (labday 136 - 145 )
- October part 2 (labday 146 - 155 )
- October part 3 (labday 156 - 166 )
- November (labday 167 - 170 )
- Cellculture
146. labday 11.10.2010
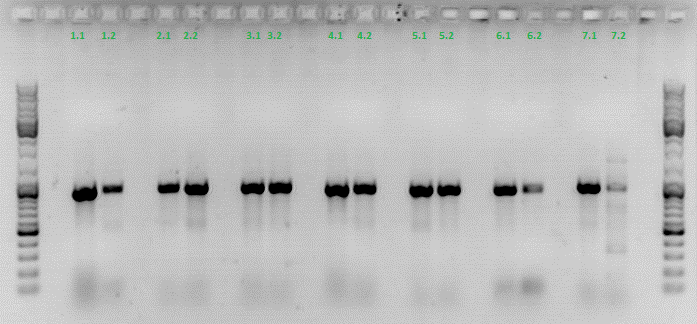
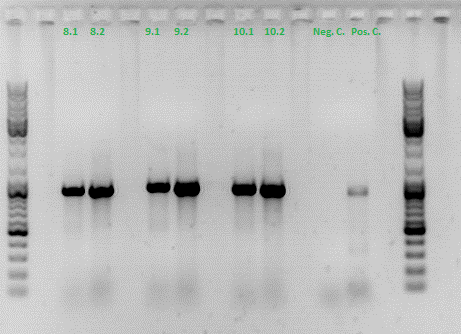
Colony PCR of Cloning VP2 Fusion and Super constructs into pSB1C3
Investigator: Achim, Hanna
Comment: Because yesterday's cloning didn't deliver a good separartion of the expected gel bands. Nevertheless ligation and trafo was performed. In order to immediately find out, whether we received successful results a colony PCR will be performed.
Two clones were picked from each plat. In addition to that a positive (pAAV_RC) and a negative control (pSB1C3_lITR) was prepared.
Used primer: 4200 rev and Cap3500 for. Expected fragment size: 885 bp.
PCR was performed following the standard protocol.
All samples match the positive control!
To do: Mini-Prep and sequencing.
Preparation of SDS-PAGE gel (10%)
Investigator: Hanna
Comment: In order to perform a Western Blot of different virus capsids (with and without capsid-motifs), 2 10% SDS polyacrylamid gels were prepared.
Resolving gel: 15 mL
- H2O: 5.9 mL
- Acryl-bisacrylamide mix (30%): 5 mL
- Tris (1.5 M, pH 8.8): 3.8 mL
- SDS (10%): 0.15 mL
- Ammonium persulfate (10%): 0.15 mL
- TEMED: 0.006 mL
Stacking gel (5%): 5 mL
- H2O: 3.4 mL
- Acryl-bisacrylamide mix (30%): 0.83 mL
- Tris (1.5 M, pH 6.8): 0.63 mL
- SDS (10%): 0.05 mL
- Ammonium persulfate (10%): 0.05 mL
- TEMED: 0.005 mL
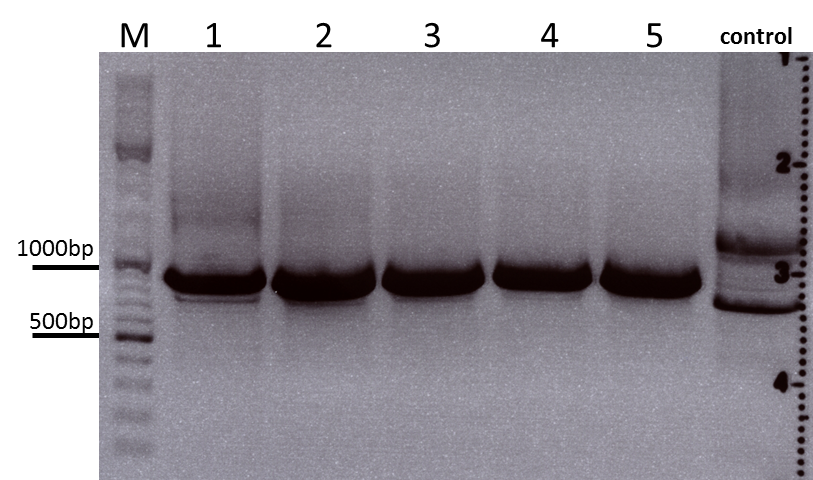
Colony PCR of p40_VP123_capins
Investigator: Bea
Comment: Since the first attempt did not work,and no cells grew on the plate and the same ligation was transformed again into BL-21 and a lot of clones grew on the plate, I decided to perform a colony PCR in order to check several colonies and to inoculate at the same day for a Midi-Prep.
Protocol:
- Primer used: O162
- Primer used: O38
The PCR products were loaded on a 1% agarose gel. The results can be seen above in the gel picture:
Result: We can see two things: The cloning of p40 to VP123 worked quiet well AND the Robust PCR Kit which was used for the first time worked as well.
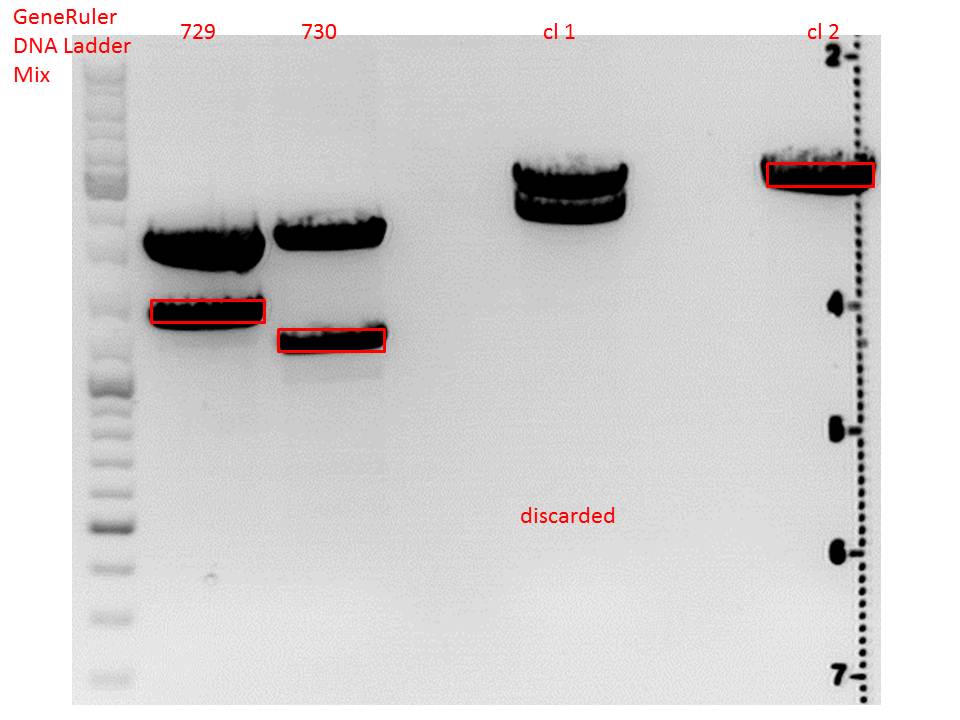
Cloning of lITR_CMV_betaglobin and lITR_phTERT_betaglobin into pSB1C3_CD
Investigator: Stefan
Comment: To produce another GOI for testing in cell culture, the cytosine deaminase needs to be assembled with lITR_promotor_betaglobin. In the next step hgH_rITR needs to be added.
Vector name:
pSB1C3_CD clone 1
pSB1C3_CD clone 2
Insert name:
pSB1C3_lITR_CMV_beta-globin (P729)
pSB1C3_lITR_phTERT_beta-globin (P730)
Digestion:
| components | volume CD clone 1 + 2 /µl | volume P729 /µl | volume P730 /µl |
| DNA | 6 | 14 | 6 |
| BSA (10x) | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 |
| Enzyme EcoI | 1 | 1 | 1 |
| Enzyme XbaI | 1 | - | - |
| Enzyme SpeI | - | 1 | 1 |
| H2O | 8 | - | 8 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 | 20 |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
CD clone 1 yielded to bands around 2500 bp to 3000 bp. Since the vector was cut only using EcoRI and SpeI, it was expected to be linearized, not to be cut into two fragments this size. Therefore, this sample was discarded and cloning was continued using CD clone 2.
Gel extraction:
Was performed according to protocol.
T4 Ligation:
| ligation name | 729 + CD cl2 | 730 + CD cl2 |
| volume of vector | 3,67 | 2,82 |
| volume of insert | 4,33 | 5,18 |
| T4 ligase buffer (10x) | 1 | 1 |
| T4 ligase | 1 | 1 |
Transformation:
Was performed according to standard protocol using BL21 cells.
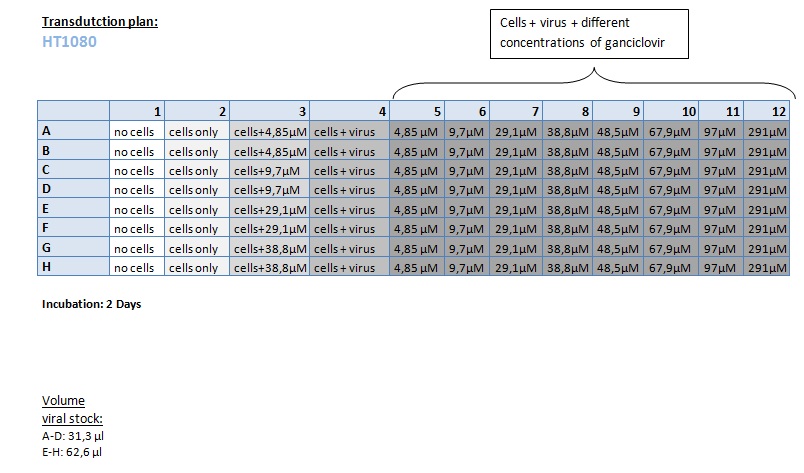
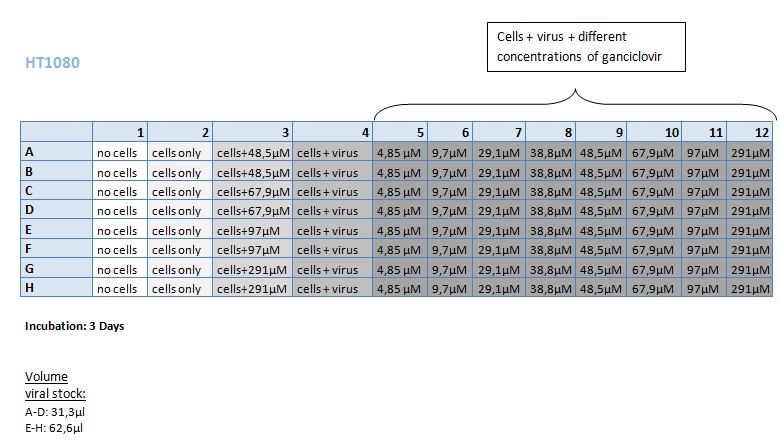
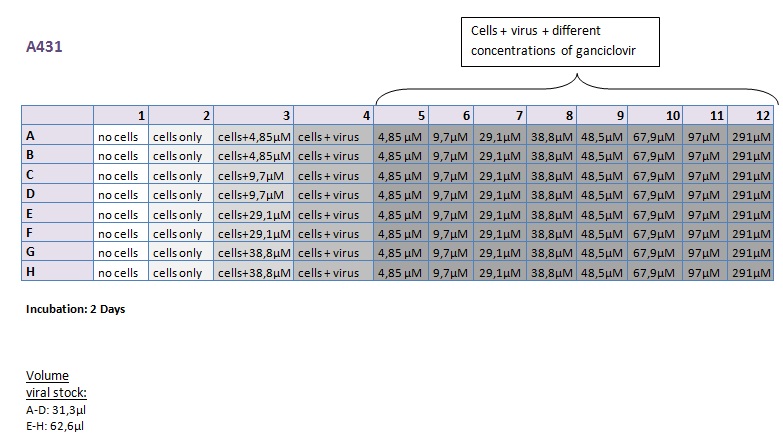
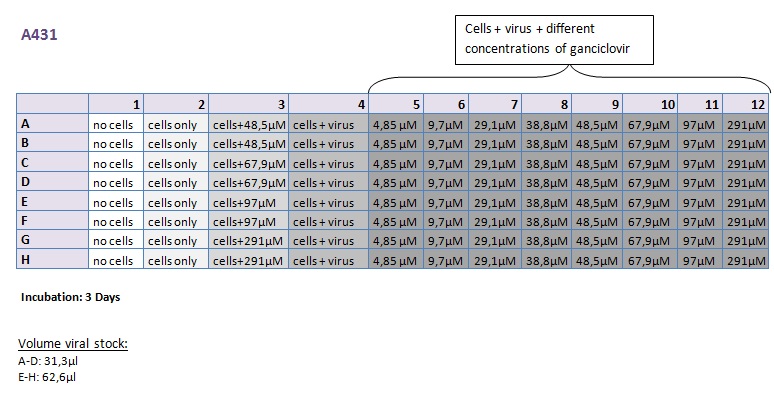
Seeding HT1080 and A431 for testing different concentrations of ganciclovir by MTT-Assay
Investigator: Kerstin, Anissa
- Seeding of 4x 96-well plates:
- Transduction plan (12.10.2010):
FACS-Analysis
Investigator: Kerstin
- Results Transduction
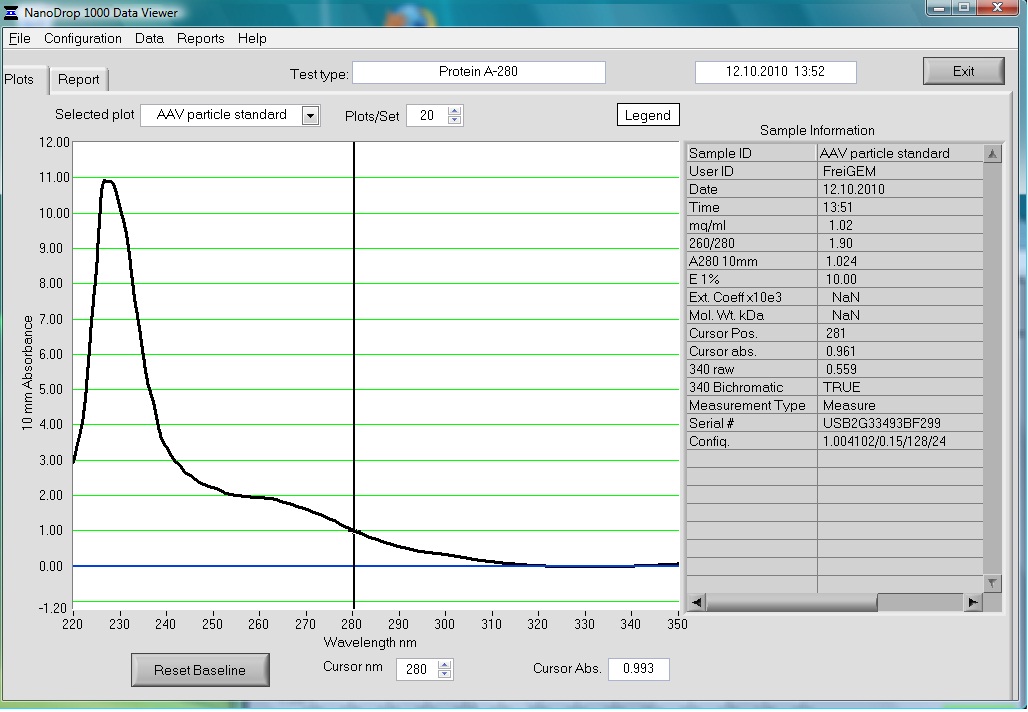
Preparation of the ELISA
Investigator: Volker
The AAV particle standard that contains 3.6x10^9 viral particles was dissolved in 500µl as described in the protocol of the Progen AAV Titration ELISA and a absorption spectrum was measured.
These purified and concentrated viral particles could be used for biophysical measurements, there for the possibility to detect the viral particles by absorption was interesting for us.
The Spectrum measured in the NanoDrop is the following:
147. labday 12.10.2010
Concentration of virus stock for Western Blot analysis
Investigator: Hanna
For the first Western Blot try, we investigate whether is is enough to concentrate the virus stock (from cell supernatant) and then to simply load it onto a SDS-PAGE gel and perfom immuno-staining.
For this purpose 8 mL virus stock were concentrated via amicon filtration.
- First, amicon filter was washed with 4 mL PBS.
- Then 4 mL virus stock was loaded onto the filter and centrifuged at 3000 rpm for 15 minutes.
- Flow-through was discarded and additional virus stock was loaded, centrifuged at 3000 rpm for 25 minutes.
- Flow-through was discarded, remaining solution was re-suspended and additional virus stock was added. Amicon filter was centrifuged at 3000 rpm for 50 minutes.
- Flow-through was discarded. Residual protein solution: 500 µL. Filter was washed with 3 mL PBS, centrifuging 35 minutes.
- 2 x 20 µL protein solution was taken and mixed with Laemmli buffer and stored over night @ 4°C.
To do: SDS-PAGE and Western Blot tomorrow.
Perfomance of another try by harvesting viruses of the cell pellet with RIPA buffer & freeze/ thaw.
Western Blot of unmodified, and modified capsids (N-terminal fusion and VP1 insertion with CFP ref. mVenus) --> size shift should be detectable.
Mini-Prep and test digestion of several constructs
Investigator: Stefan
Glycerol stocks were prepared:
- B651 = pSB1C3_P40_VP123
- B652 = pSB1C3_litr_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR
- B653 = pSB1C3_litr_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR
- B654 = pSB1C3_Iitr_CMV_ß-globin_mGMK_TK30_hgH_rITR
- B655 = pSB1C3_Iitr_CMV_ß-globin_mGMK_TK30_hgH_rITR
- B656 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO
- B657 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO
- B658 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO
- B659 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO
- B660 = pSB1C3_litr_phTERT_ß-globin_cfp_hgH-ritr
- B661 = pSB1C3_litr_phTERT_ß-globin_cfp_hgH-ritr
Mini-Prep was performed according to standard protocol:
- P821 = pSB1C3_P40_VP123 c = 327,4 ng/µl
- P822 = pSB1C3_litr_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR c = 342,9 ng/µl
- P823 = pSB1C3_litr_phTERT_ß-globin_mGMK_TK30_pSTI_SDM_hgH_rITR c = 324,0 ng/µl
- P824 = pSB1C3_Iitr_CMV_ß-globin_mGMK_TK30_hgH_rITR c = 81,6 ng/µl
- P825 = pSB1C3_Iitr_CMV_ß-globin_mGMK_TK30_hgH_rITR c = 148,4 ng/µl
- P826 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO c = 105,8 ng/µl
- P827 = pSB1C3_CMV_VP123_453_Z34C_HSPG-KO c = 121,1 ng/µl
- P828 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO c = 188,4 ng/µl
- P829 = pSB1C3_CMV_VP123_453_RGD_HSPG-KO c = 140,4 ng/µl
- P830 = pSB1C3_litr_phTERT_ß-globin_cfp_hgH-ritr c = 197,2 ng/µl
- P831 = pSB1C3_litr_phTERT_ß-globin_cfp_hgH-ritr c = 207,3 ng/µl
Test digestion:
| Components | P803 + P804 / µl |
| DNA | 2,5 |
| Buffer 4 | 1 |
| BSA (10x) | 1 |
| PstI | 0,3 |
| H2O | 5,2 |
| Total volume | 10 |
Gel:
0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes
Comment: .
148. labday 13.10.2010
149. labday 14.10.2010
150. labday 15.10.2010
151. labday 16.10.2010
152. labday 17.10.2010
153. labday 18.10.2010
154. labday 19.10.2010
155. labday 20.10.2010
 "
"