Team:Alberta/PartsDesigner
From 2010.igem.org
The Plasmid Designer
The plasmid designer gives students and researchers the ability to design and validate their plasmids in in silico. Through an intuitive drag and drop interface, users can piece their plasmids together BioByte by BioByte. During this, the program provides feedback. It can check whether the desired plasmid will circularize properly. The presence of a selectable marker and an origin of replication is also checked. The user can see the resulting annotated sequence of the plasmid as it is being built. If the plasmid is being designed for an experiment, the software will generate a protocol for producing that plasmid given materials available in the kit, and insert that protocol into the user's online lab manual.
Part Graphics
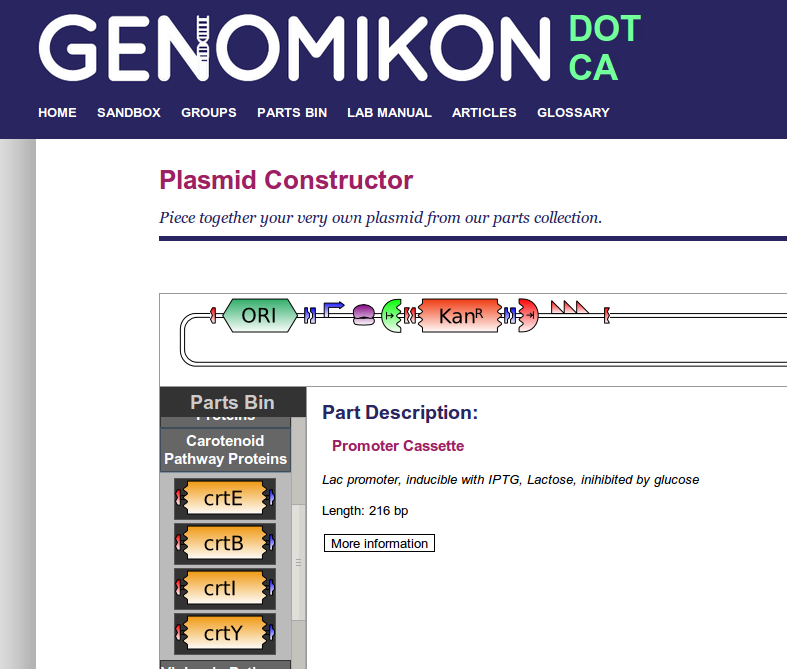
In order to make the interface as intuitive as possible, icon graphics were chosen that would provide clear visual feedback of the key structures and functional sequences present on the plasmid.
The A and B Ends
'A' sticky ends are denoted as two interlocking red pieces, and 'B' sticky ends are denoted as two differently interlocking blue pieces. This gives a quick reminder that 'AB' parts can only be stuck to 'BA' parts in sequence, and vice versa.
Origins of Replication
Origins of replication are given a unique block shape with the symbol, 'ORI' written on them.
Open Reading Frames
Open reading frames are blocks of various colors with a short acronym or abbreviation of its name on them. The ends are given a 'torn away' appearance that interlocks with the symbols for start and stop codons. This is to remind the user that BioByte open reading frame parts do not contain a start or stop codon. Therefore, the user must remember to proceed an ORF BioByte with a start codon, and include a stop codon after. Otherwise, interesting gene fusions could be created, intentionally or not.
Selectable Markers
Selectable markers are sometimes simply a special case of ORF, with no start or stop codon. Others are full cassettes, which contain promoters, start and stop codons, and anything else necessary for expression. This distinction is made clear with appropriate graphics.
Promoters
Transcriptional promoters are indicated with a bent arrow.
Ribosome Binding Sites
Ribosome binding sites are indicated with a very stylized ribosome-like blob shape.
Start and Stop Codons
Start and stop codons are indicated with a half-circle, with a 'torn' appearance such that they appear to interlock neatly with the open reading frame icons. Start codons are green, and have an arrow extending right. Stop codons are red, with an arrow ending at a vertical line.
Transcriptional Terminators
Transcriptional terminators are visualized by three successive red sawtooth shapes.
How It Works Together
If one wanted to build a simple, viable plasmid that could replicate and provide antibiotic resistance, one would need to drag together the following parts from the parts bin:
- An 'AB' origin of replication BioByte
- A 'BA' promoter, ribosome binding site, start codon BioByte
- An 'AB' ampicillin resistance open reading frame BioByte
- A 'BA' stop codon, transcriptional terminator BioByte
In the plasmid designer, it looks like this:
Sequence View
As soon as parts are dragged into the plasmid, the annotated sequence is shown to the user, and is updated in real time.
 "
"