Team:RMIT Australia/Modeling
From 2010.igem.org


Modelling
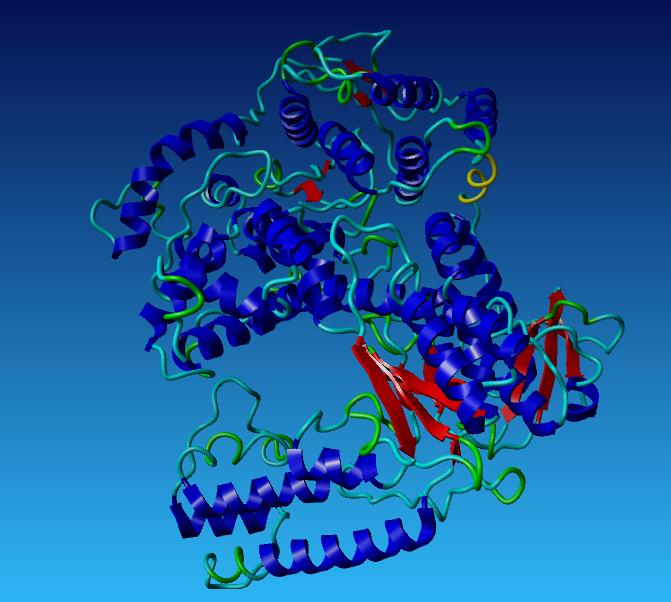
Taq DNA Polymerase
Taq Polymerase with no mutations
Aliphatic Index: 98.33
Ext Coefficient: 112760
Theoretical PI: 6.04
Instability Index: 32.93
We redesigned TAQ so it is suitable to undergo ligation independent cloning, to do this we changed the N and C terminal.
This is the redesigned Taq Polymerase
Aliphatic Index: 96.46
Ext Coefficient: 107260
Theoretical PI: 6.09
Instability Index: 33.39
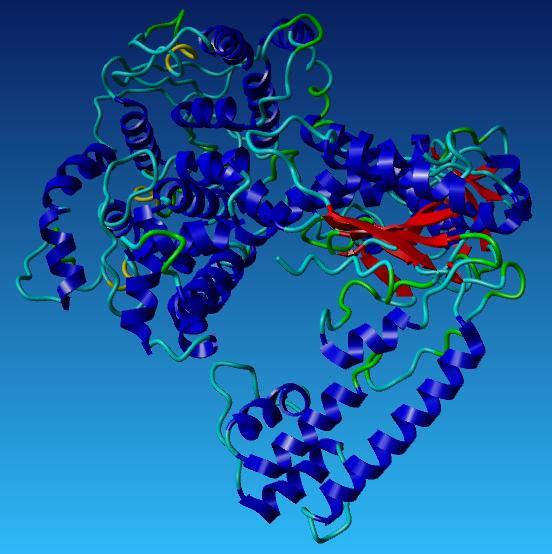
The wild type Taq Polymerase was changed at the N and C terminal, with the Root Mean Square Deviation of 6.8834. This number indicates the measure of the average distance between the backbones of the two superimposed proteins.
The picture below shows the differences between the wild type Taq Polymerase and the redesigned Taq at the N and C terminus.
Our Taq Polymerase with mutations
485 Asparagine - Valine
515 Serine - Leucine
540 Lysine - Leucine
659 Arginine - Isolucine
663 Lysine - Leucine
Fold-it

Foldit is a revolutionary new computer game enabling you to contribute to important scientific research. This page describes the science behind Foldit and how your playing can help.
[http://fold.it/portal/ Click Here]
 "
"