Team:Calgary/12 August 2010
From 2010.igem.org

Notebook
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
Thursday August 12, 2010
Chris
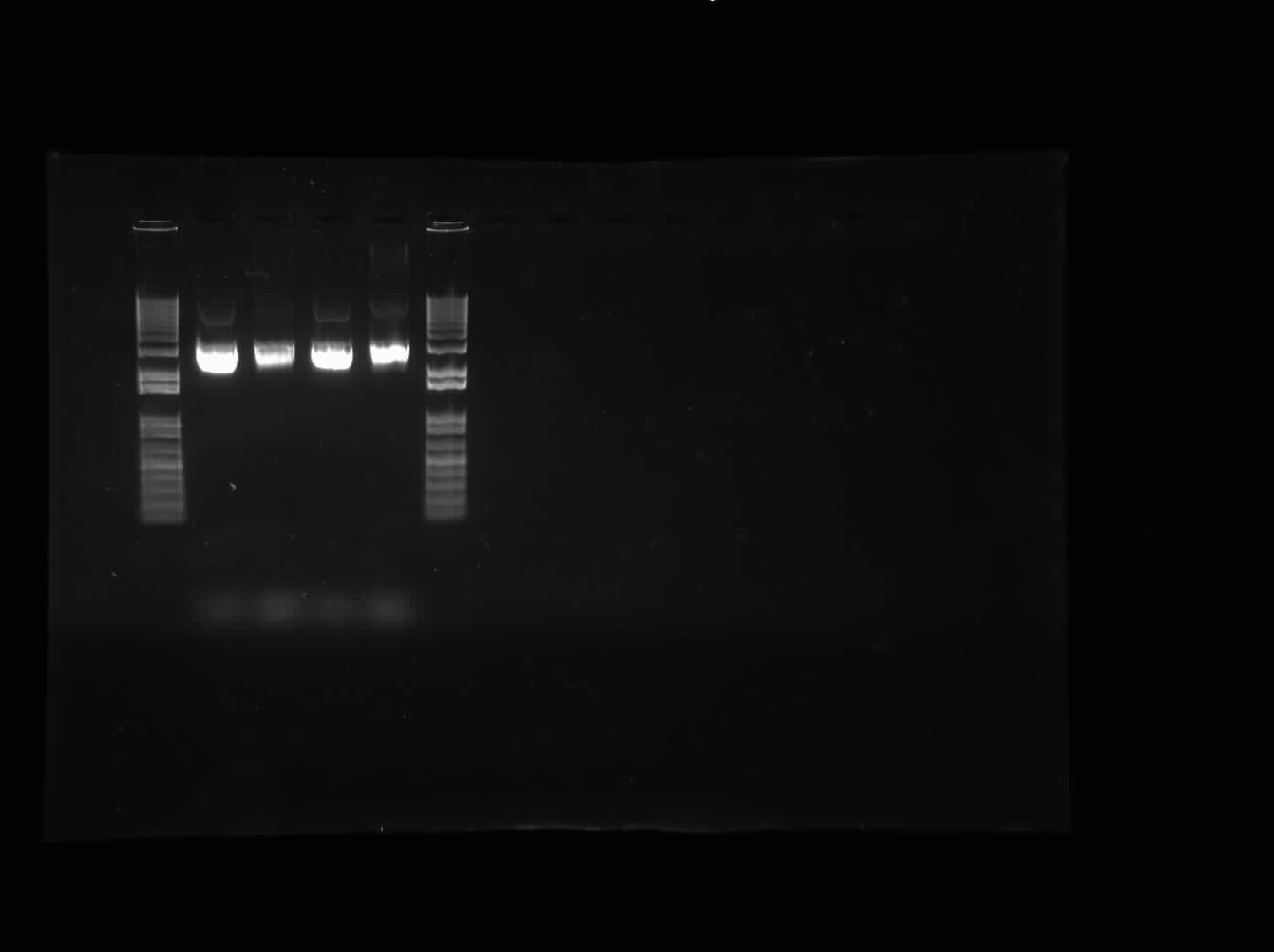
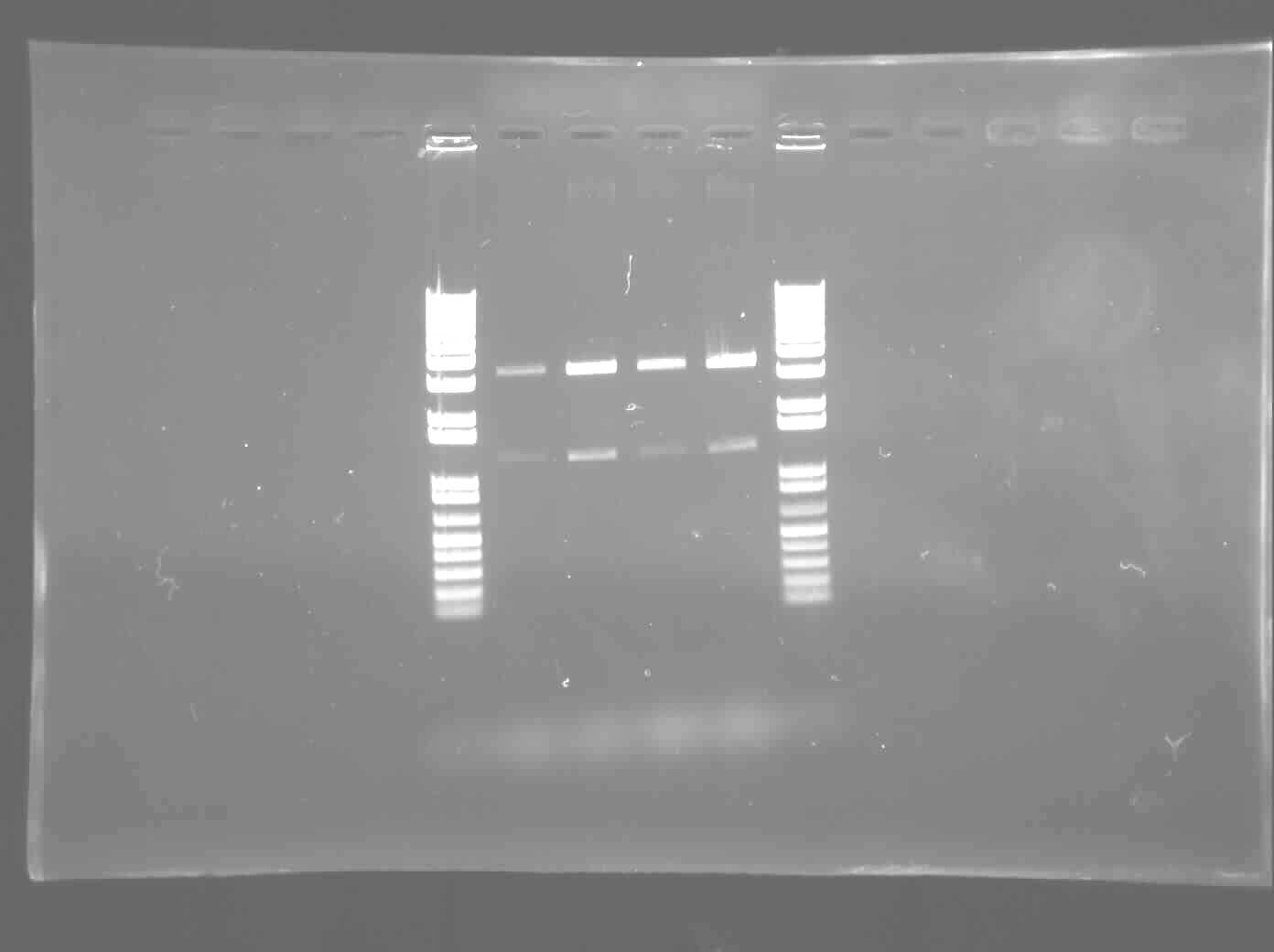
Today, I started with a ton of plasmid preparations with I13504, iPBAB, I0500-I13504, and I13507 colonies. The concentrations are once again, available upon demand. Then, I began a digest of ibpAB to with the enzymes EcoRI and PstI to insert it into an AK Biobrick vector. I also digested the CpxP promoter that was inserted into AK plasmid with EcoRI and PstI. The results of the gel with the digested CpxP and ibpAB are shown on the first gel on the right. The second gel was the ibpAB promoter uncut in original state. Finally, I set up another digest for ibpAB to run with T4 DNA Ligase tomorrow as we are out of Quick Ligase.
Raida
Today I ran the gel electrophoresis of the restriction digest that Emily did on Thursday. The results are positive, the bands are at the right place : 1.2 and 3.9 kb. These will be sent down for sequencing tomorrow. I also ran a pcr of malEssdel and malE31ssdel to insert them in TOPO vector.
I also worked on Ethics paper. I completed the Environment and synthetic biology portion of the paper.
Himika
Today I read more papers regarding inclusion body formation and modeling the same in SImbiology.
 "
"