Team:TU Delft/2 August 2010 content
From 2010.igem.org
Alkane Sensing, Solvent Tolerance and Salt Tolerance
by Pieter
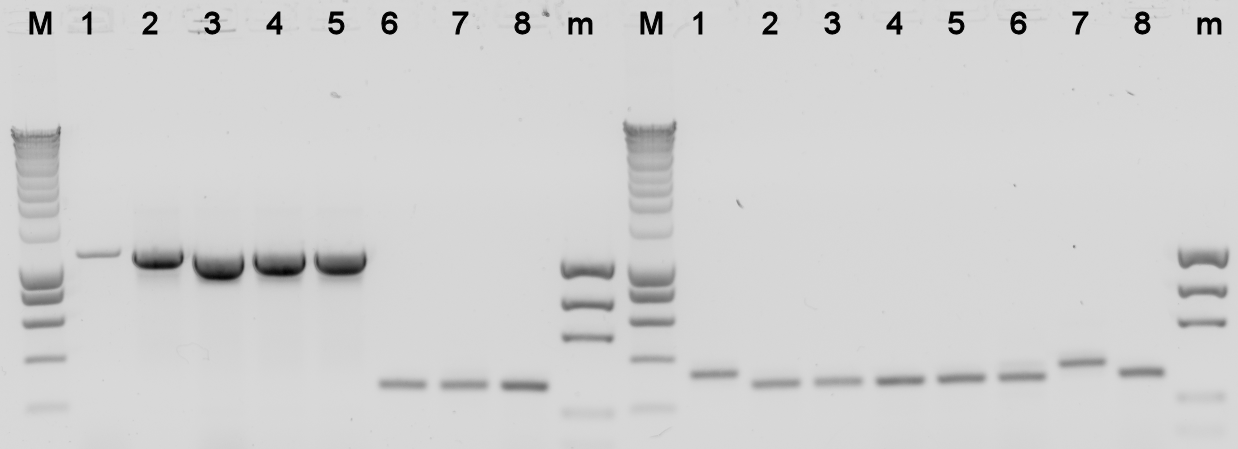
The plates containing yesterday's ligations contained colonies, to check whether they really contain the desired BioBrick a colony PCR was done, and the used colonies were grown in liquid LB medium over night. The results from the PCR were analysed on a 1% agarose gel.
- BBa_K398305 = Alks -> E0240
- BBa_K398101 = bbc1 -> J61101
- BBa_K398402 = PhPFDalpha -> J61101
- BBa_K398403 = PhPFDbeta -> J61101
Lane Description:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| M | SmartLadder | n/a | n/a | n/a | |
| 1 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 2 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 3 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 4 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 5 | BBa_K398305 | 3772 | G00101 + G00100 | X | |
| 6 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| 7 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| 8 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| m | EZ Ladder | n/a | n/a | n/a | |
| M | SmartLadder | n/a | n/a | n/a | |
| 1 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| 2 | BBa_K398101 | 1007 | G00101 + G00100 | X | |
| 3 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 4 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 5 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 6 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 7 | BBa_K398402 | 836 | G00101 + G00100 | X | |
| 8 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| m | EZ Ladder | n/a | n/a | n/a |
Lane Description:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| M | SmartLadder | n/a | n/a | n/a | |
| 1 - 7 | Eva's samples | n/a | n/a | n/a | |
| m | EZ tLadder | n/a | n/a | n/a | |
| 1 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| 2 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| 3 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| 4 | BBa_K398403 | 734 | G00101 + G00100 | X | |
| M | SmartLadder | n/a | n/a | n/a |
Emulsifier
When going over the plates in the fridge we noticed a few white colonies on plates from 13-07-'10. Previously we thought there were only red colonies. So we decided to do a colony PCR on all 12 white colonies.
Lane Description:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| M | SmartLadder | n/a | n/a | n/a | |
| 1 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 2 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 3 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 4 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 5 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 6 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 7 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 8 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 9 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 10 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 11 | BBa_K398202 | 392 | G00101 + G00100 | ||
| 12 | BBa_K398202 | 392 | G00101 + G00100 |
Alkane degradation
For the next step in BioBrick formation a digestion was done:
| # | Sample | Enzyme 1 | Enzyme 2 | Enzyme 3 | Buffer | BSA | Needed fragment |
| 1 | 1 μg 007T | EcoRI | SpeI | BamH1 | 2 (Biolabs) | ✓ | ‘E – J61100-AlkB2 – S’ |
| 2 | 1 μg 008A | EcoRI | XbaI | 2 (Biolabs) | ✓ | ‘E – J61100-rubA3 – X’ | |
| 3 | 1 μg 010A | EcoRI | SpeI | AseI | 2 (Biolabs) | ✓ | ‘E – J61100-rubR – S’ |
| 4 | 2 μg B0015 | EcoRI | XbaI | 2 (Biolabs) | ✓ | ‘E – B0015 – pSB1AK3 – X’ | |
| 5 | 1 μg 017A | EcoRI | SpeI | AseI | 2 (Biolabs) | ✓ | ‘E – J61100-ladA – S’ |
| 6 | 1 μg 018A | XbaI | PstI | AseI | 2 (Biolabs) | ✓ | ‘X – J61101-ADH – P’ |
| 7 | 1 μg 019A | EcoRI | SpeI | AseI | 2 (Biolabs) | ✓ | ‘E – J61107-ALDH – S’ |
| 8 | 1 μg pSB1K3 | EcoRI | PstI | 2 (Biolabs) | ✓ | ‘E – pSB1K3 – P’ |
Results of the digestion on 1% agarose gel
The gel was runned at 100V for 1 hour, loaded 5 μL of marker and 5 μL sample + 1 μL loadingbuffer
Lane description:
| # | Description | Expected Length (bp) | Status |
| 1 | Smartladder | ||
| 2 | ‘E – J61100-AlkB2 – S’ (007T) | ✗ | |
| 3 | Undigested 007T | ✗ | |
| 4 | ‘E – J61100-rubA3 – X’ (008A) | ✓ | |
| 5 | Undigested 008A | ✓ | |
| 6 | ‘E – J61100-rubR – S’ (010A) | ✓ | |
| 7 | Undigested 010A | ✓ | |
| 8 | ‘E – B0015 – pSB1AK3 – X’ | ✓ | |
| 9 | pSB1AK3-B0015 | ✓ | |
| 10 | ‘E – J61100-ladA – S’ (017A) | ✓ | |
| 11 | Undigested 017A | ✓ | |
| 12 | ‘X – J61101-ADH – P’ (018A) | ✓ | |
| 13 | Undigested 018A | ✓ | |
| 14 | ‘E – J61107-ALDH – S’ (019A) | ✓ | |
| 15 | Undigested (019A) | ✓ | |
| 16 | ‘E – pSB1K3 – P’ | ✓ | |
| 17 | Smartladder |
 "
"