Team:Calgary/26 July 2010
From 2010.igem.org

Monday July 26, 2010
Jeremy
To do: I13504 K239000 Miniprep C3, I13507 K239000 Miniprep all, I13504 K135000 Redo, I13507 K135000 I13504 K239000(plasmid switch) and PCR, I13507+K135000 C3,C4,C6.
PCR sequencing -order primers, gradient PCR
Dev
Today, I worked on the construction of K135000 with I13504.
Chris
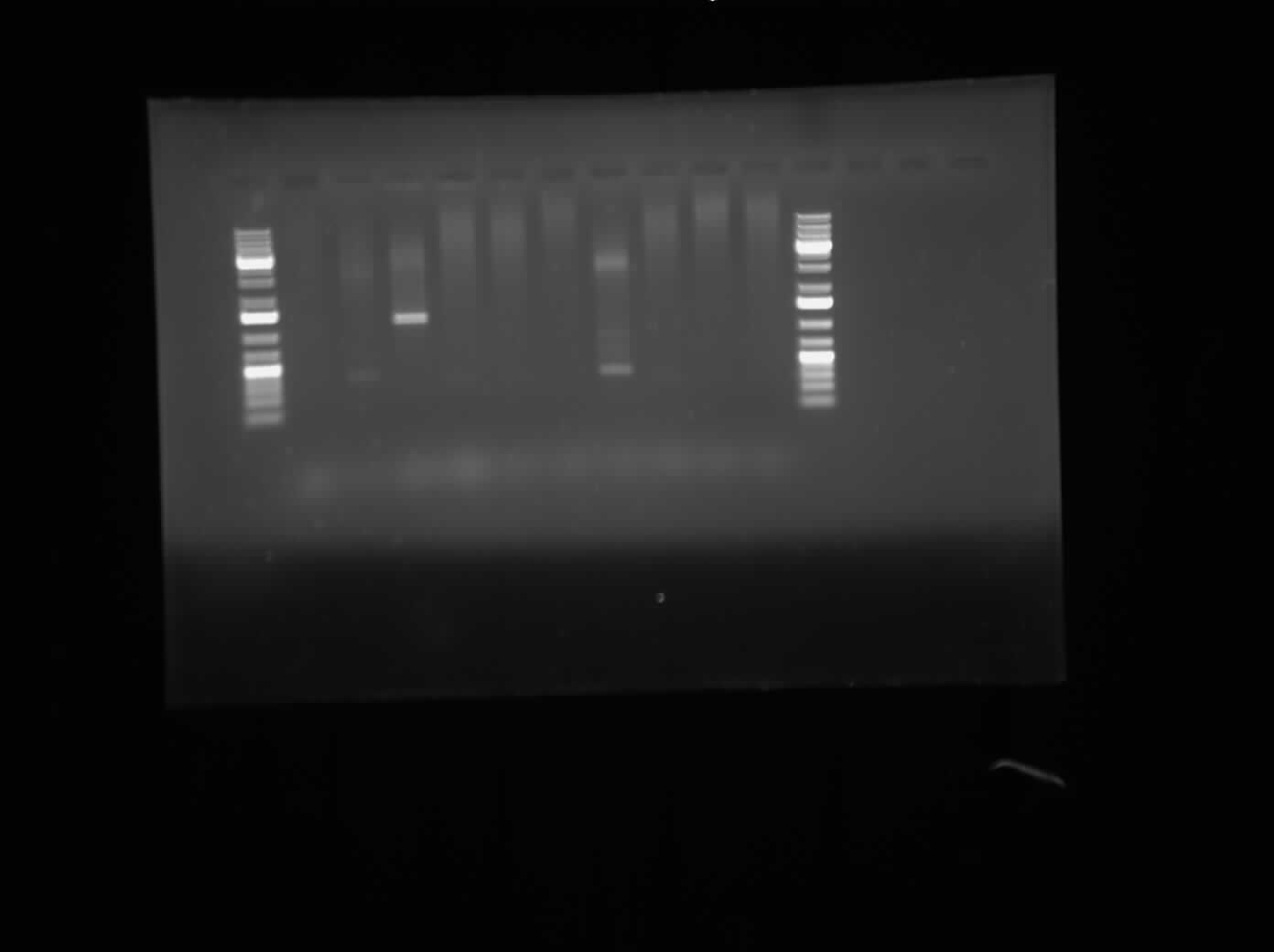
Today, I ran agarose gel electrophoresis of the different CpxP promoters that were put into Ampicillin-chloramphenicol and ampicillin-kanamycin plasmids. The first gel that was run was of the promoters having been digested with the restriction enzymes XbaI and PstI. This gel can be seen on the right. The result of the gel showed bands around approximately 3000 bp, meaning tht the plasmid was not cut by the enzymes. The second gel that was run of the CpxP promoter after a regular PCR with Biobrick primers. The bands were very inconclusive and thus, we cannot conclude that the CpxP promoter is present.
After the bands showed up inconclusive, I suspect that I may have forgotten to add an ingredient to the PCR Master Mix so I will redo this overnight. In the meantime, I am restriction digesting the parts of I13507 (RFP Construct), I13504 (GFP construct), K239000 (DegP) and K135000 (CpxR). The restriction enzymes used are XbaI and PstI for the two constructs and SpeI and PstI for the CpxR and DegP promoters. They will be allowed to digest overnight and then will be ligated in the morning.
I also contacted Jennifer Logan, our Health Sciences coordinator at the University of Calgary, about setting up an account with IDT DNA Technologies.
Raida
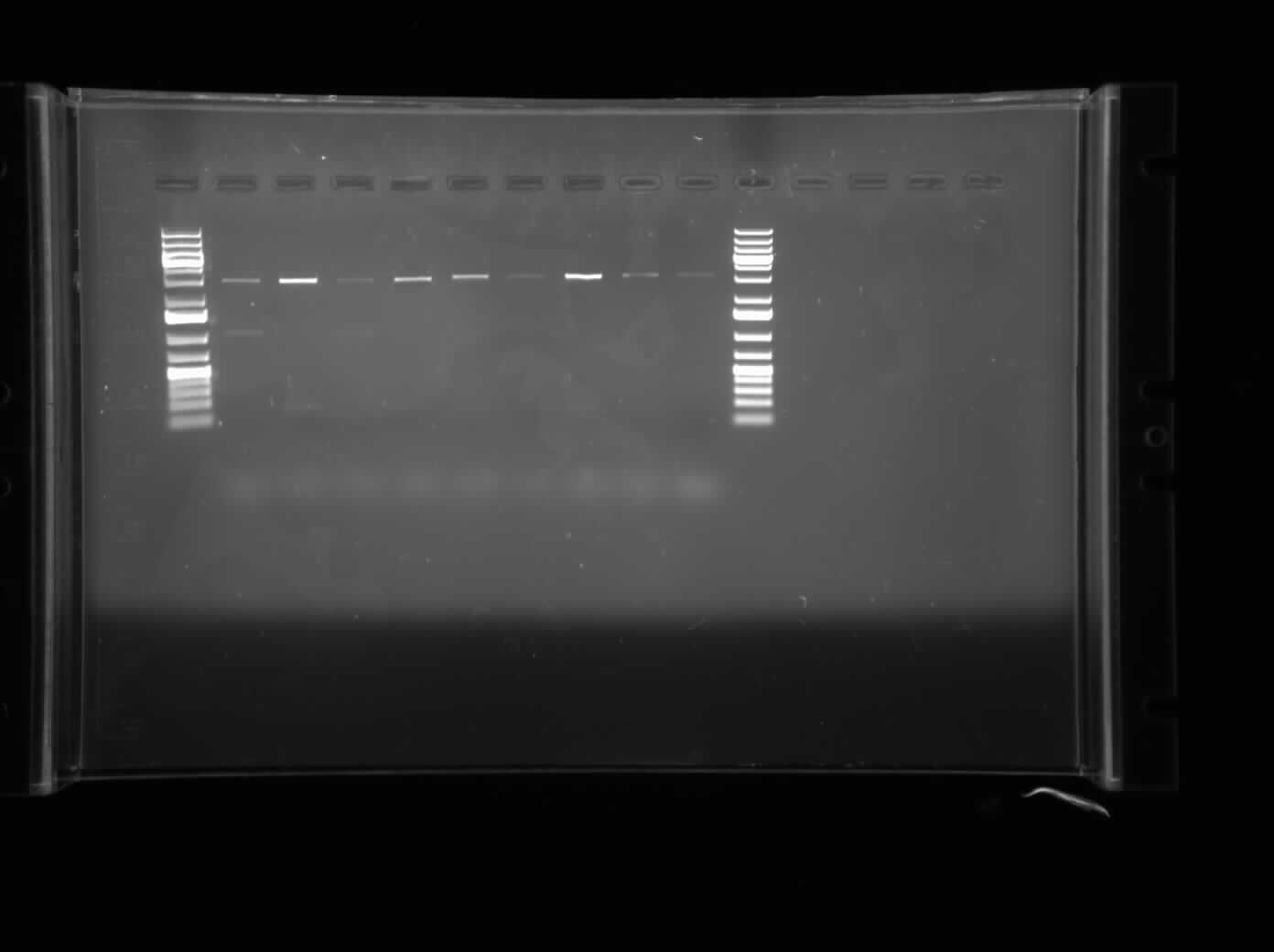
Today I ran an agarose gel electrophoresis of the PCR product containing the diluted MalE Gene that was amplified on June 23, 2010 PCR. The primers used for this particular PCR were the ones containing the internal restriction sites. Hence, a successful annealing would be indicated by bands containing the restriction sites. However, the gel showed no bands, hence no annealing of the BBK-primers with the MalE Gene.
Today I'm also reading on the Ethics Paper from 2009 Calgary iGEM competition and am trying to find two goals for this year's paper.
Emily
Today I diluted the BBK-CP primers (enough for the rest of eternity I hope) as well as the malEBBKdelSS primer, which is going to be used to Biobrick the malE and malE31 genes that have their signal sequences deleted. I also spent some time looking up the details of the plasmids that we have been shipped from the Raivio lab in Edmonton in order to determine what kind of tests we will be using them for when our system is built. I also met with Leanne Y. on main campus. She is the media relations coordinator for the Faculty of Science. We talked about ways to get some more publicity for our iGEM team this year both during the Summer, as well as closer ot the two jamborees in the Fall. Finally I spent some time coming up with a Wetlab plan for the rest of the Summer.
Patrick
We decided to change our modelling project, hopefully for the last time. We are no longer doing plasmid copy number because its applicability to solving other problems seems to be quite limited. Instead, we are now looking at the factors that affect the formation of inclusion bodies, which are clumps of aggregate proteins (i.e. proteins that misfold). I am now looking into these factors and debating which ones would be the best candidate for modelling.
No notebook page exists for this date. Sorry! "
"