Team:Newcastle/9 August 2010
From 2010.igem.org

| |||||||||||||
| |||||||||||||
Contents |
Amplification of Pspac_oid promoter by PCR
Aim
The aim of this experiment is to amplify the Pspac_oid promoter fragment from plasmid pMutin4 for the construction of rocF BioBrick using Phusion PCR.
Materials and Protocol
Please refer to PCR for Phusion PCR protocol. The details for the 4 PCR reactions are mentioned below:
PCR
| Tube | Part to be amplified | DNA fragment consisting the part | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time* (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | Pspacoid Promoter | pMutin4 | P1P1 forward | P2P1 reverse | 58 | 106 approx. | 15 |
| 2 | Pspacoid Promoter | pMutin4 | P1P1 forward | P2P1 reverse | 59 | 106 approx. | 15 |
Table 1: Table represents 4 different Phusion PCR reactions for the amplification of Pspac_oid promoter, so that it can be ligated together with other fragments for the construction of rocF with the help of Gibson Cloning method.
- The extension rate of the Phusion polymerase is 1Kb/ 30 seconds. Therefore the extension time of each PCR reaction is different.
- To learn more about the rocF fragments, please refer to the Cloning strategy for rocF.
Discussion
For the gel electrophoresis results, please refer to the bottom section.
Amplification of Pspac_oid promoter by PCR
Aim
The aim of this experiment is to amplify the Pspac_oid promoter fragment from plasmid pMK-RQ containing Biobrick kinA and plasmid pMK-RQ containing stochastic switch developed by Team Newcastle 2009 for the construction of rocF BioBrick using Phusion PCR.
Materials and Protocol
Please refer to PCR for Phusion PCR protocol. The details for the 2 PCR reactions are mentioned below:
PCR
| Tube | Part to be amplified | DNA fragment consisting the part | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time* (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | Pspacoid Promoter | Plasmid containing kinA | P1P1 forward | P2P1 reverse | 58 | 106 approx. | 15 |
| 2 | Pspacoid Promoter | Plasmid containing kinA | P1P1 forward | P2P1 reverse | 59 | 106 approx. | 15 |
| 3 | Pspacoid Promoter | Plasmid containing stochastic switch | P1P1 forward | P2P1 reverse | 58 | 106 approx. | 15 |
| 1 | Pspacoid Promoter | Plasmid containing stochastic switch | P1P1 forward | P2P1 reverse | 59 | 106 approx. | 15 |
Table 2: Table represents 2 different Phusion PCR reactions for the amplification of Pspac_oid promoter, so that it can be ligated together with other fragments for the construction of rocF with the help of Gibson Cloning method.
- The extension rate of the Phusion polymerase is 1Kb/ 30 seconds. Therefore the extension time of each PCR reaction is different.
- To learn more about the rocF fragments, please refer to the Cloning strategy for rocF.
Discussion
For the gel electrophoresis results, please refer to the bottom section.
Gel Electrophoresis for the amplified Pspac_oid promoter
Aim
The aim of the experiment is to perform gel electrophoresis for the two PCR reactions Pspac_oid promoter amplified from plasmid pMutin4 and from plasmid pMK-RQ containing Biobrick kinA and plasmid pMK-RQ containing stochastic switch developed by Team Newcastle 2009.
Materials and Protocol
Please refer to: Gel electrophoresis.
Result
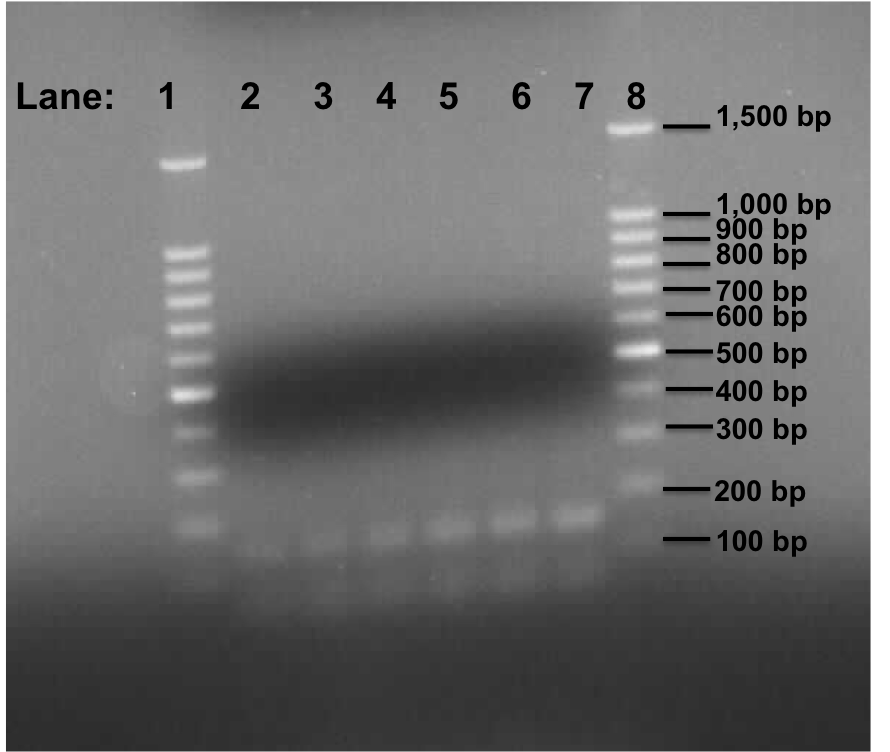
Figure 1: Gel electrophoresis of the lacI and Pspac_oid promoter.
- Lane 1: 100bp DNA ladder
- Lane 2: Plamid pMutin4 containing Pspac_oid promoter
- Lane 3: Plamid pMutin4 containing Pspac_oid promoter
- Lane 4: Plamid pMK-RQ (kinA BioBrick) containing Pspac_oid promoter
- Lane 5: Plamid pMK-RQ (kinA BioBrick) containing Pspac_oid promoter
- Lane 6: Plamid pMK-RO (stchastic switch BioBrick) containing Pspac_oid promoter
- Lane 7: Plamid pMK-RO (stchastic switch BioBrick) containing Pspac_oid promoter
- Lane 8: 100bp DNA ladder
| Pspac_oid pormoter | |
|---|---|
| Size of the Fragment (in bp) | 148 approx. |
Table 3: Table represents the size of the Pspac_oid fragment represented as bands on the gel in all the lanes.
Discussion
Two bands were found in all the lanes which is of approximately 150 bp and 80 in size.
Conclusion
This experiment shows that the PCR reaction was not successful for the Pspac_oid fragment. The 80 bp band might be primer dimers. When the primer sequence was checked again, we found that there is a high complementarity within the primers itself and thus the amplification of the Pspac_oid fragment is not taking place due to the primer pairs joining together. The 150 bp band could be that of the Pspac_oid fragment. Therefore we will be doing gel extraction tomorrow to confirm if the 150 bp band is the Pspac_oid promoter.
 
|
 "
"